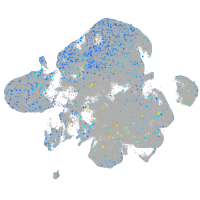
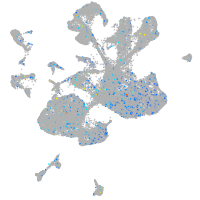
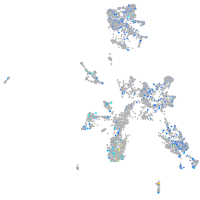
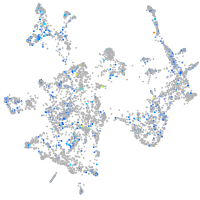
NFS1 cysteine desulfurase
ZFIN 
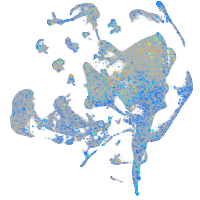
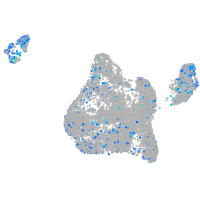
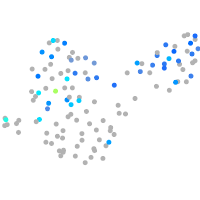
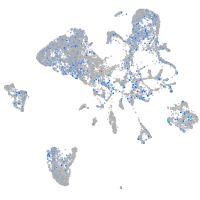
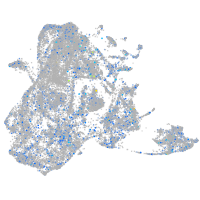
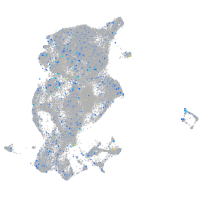
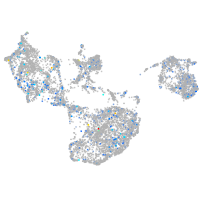
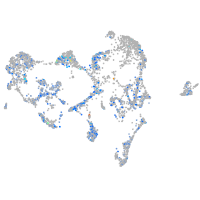
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| id1 | 0.072 | stmn1b | -0.053 |
| ranbp1 | 0.060 | elavl3 | -0.052 |
| eif5a2 | 0.059 | rtn1a | -0.051 |
| mki67 | 0.059 | gpm6aa | -0.044 |
| tuba8l | 0.058 | gng3 | -0.043 |
| zfp36l1a | 0.058 | gpm6ab | -0.042 |
| pa2g4b | 0.058 | ckbb | -0.041 |
| hmgb2a | 0.058 | ptmaa | -0.041 |
| pcna | 0.057 | tmsb | -0.040 |
| mcm7 | 0.056 | rtn1b | -0.038 |
| snu13b | 0.055 | sncb | -0.036 |
| cnbpa | 0.055 | atp6v0cb | -0.036 |
| tuba8l4 | 0.055 | sox4a | -0.035 |
| dut | 0.055 | zc4h2 | -0.034 |
| nhp2 | 0.055 | fez1 | -0.034 |
| nop10 | 0.054 | rnasekb | -0.034 |
| boc | 0.054 | tuba1c | -0.033 |
| sdc4 | 0.054 | pvalb1 | -0.033 |
| anp32b | 0.054 | gng2 | -0.033 |
| msna | 0.054 | si:dkeyp-75h12.5 | -0.031 |
| her2 | 0.053 | mllt11 | -0.031 |
| ccnd1 | 0.053 | pvalb2 | -0.031 |
| nutf2l | 0.053 | gap43 | -0.031 |
| nop58 | 0.053 | vamp2 | -0.031 |
| zgc:56493 | 0.052 | stmn2a | -0.030 |
| msi1 | 0.052 | id4 | -0.030 |
| rrm1 | 0.052 | marcksl1b | -0.030 |
| cad | 0.052 | olfm1b | -0.030 |
| npm1a | 0.052 | elavl4 | -0.030 |
| paics | 0.052 | actc1b | -0.029 |
| lbr | 0.052 | ywhag2 | -0.029 |
| ccng1 | 0.052 | tubb5 | -0.029 |
| dkc1 | 0.052 | dpysl3 | -0.029 |
| seta | 0.051 | kdm6bb | -0.029 |
| mdka | 0.051 | myt1b | -0.029 |