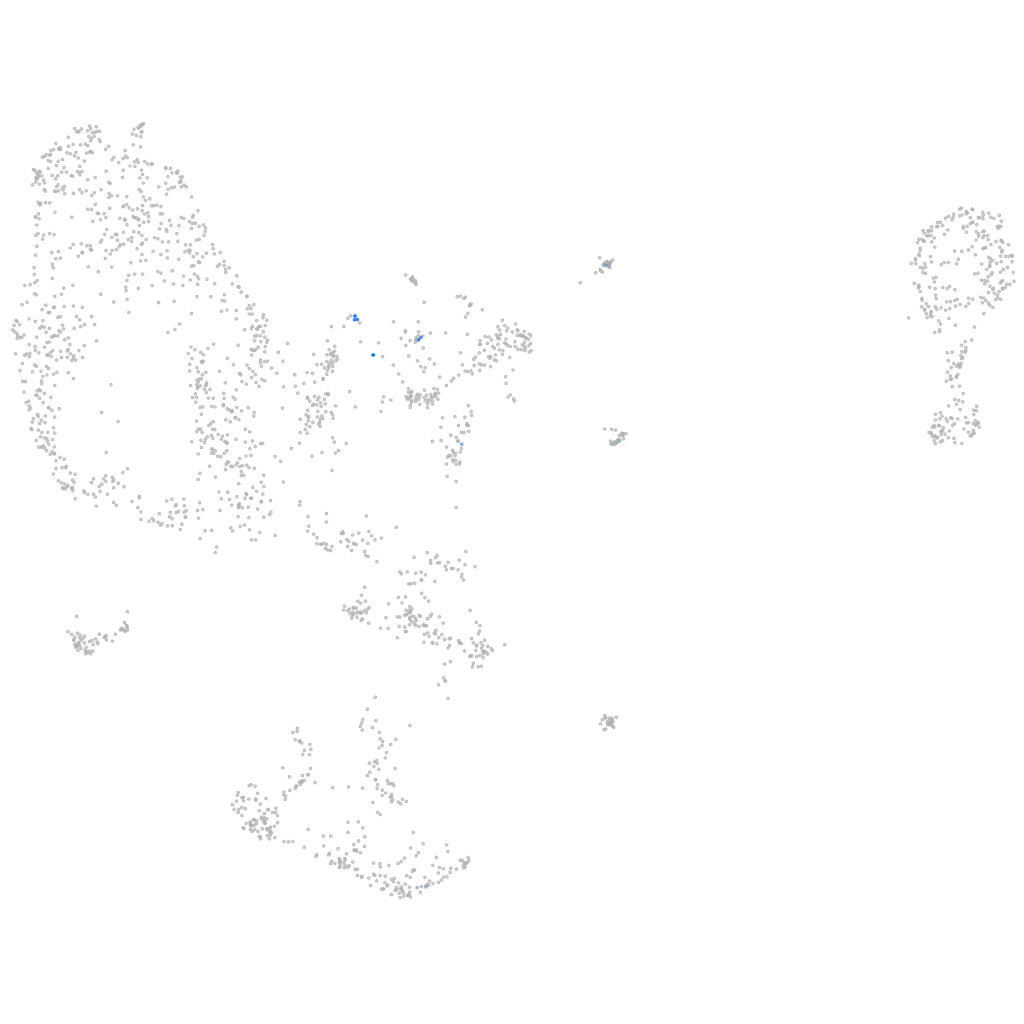
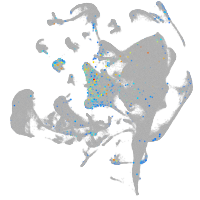
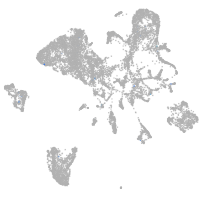
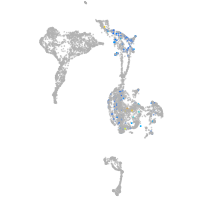
nuclear factor of activated T cells 1
ZFIN 
Expression by stage/cluster


Correlated gene expression