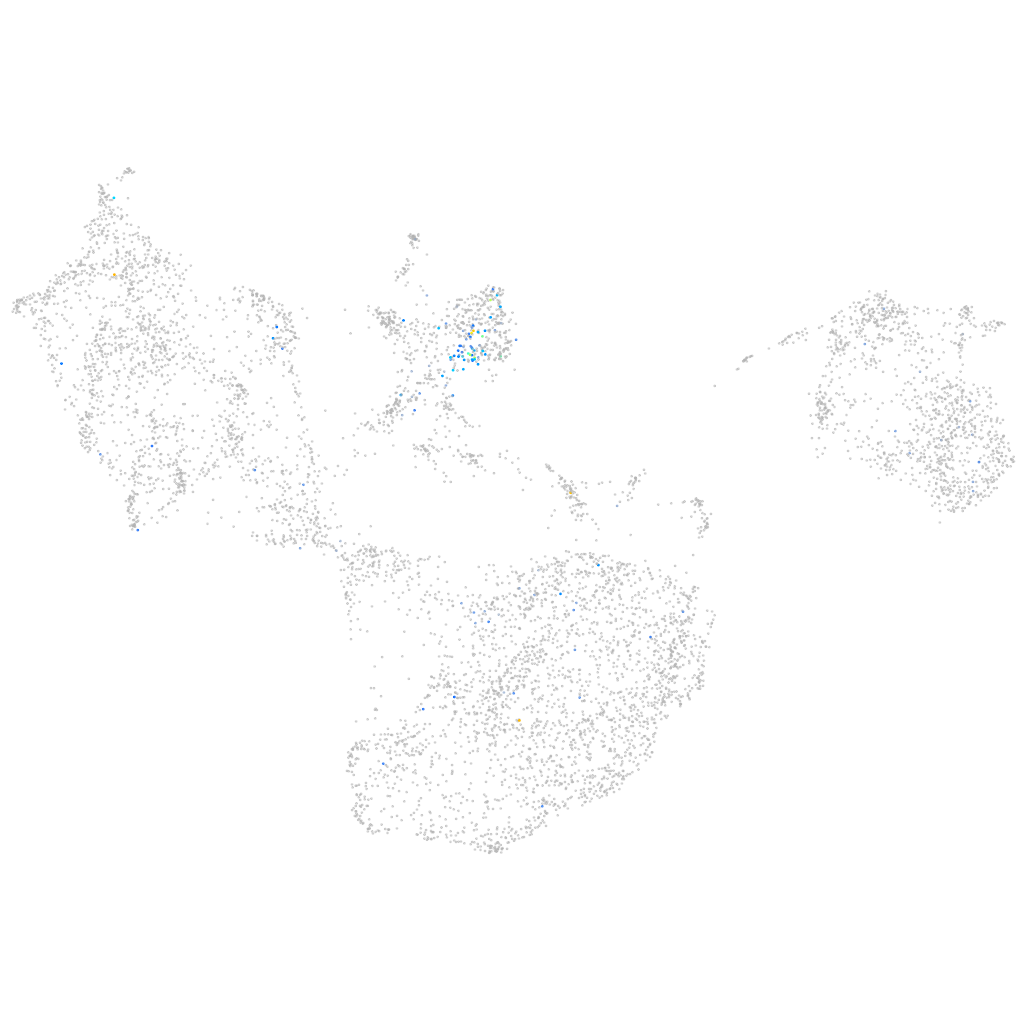
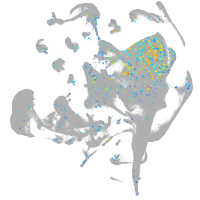
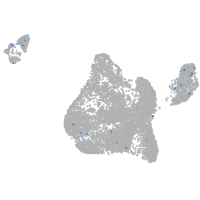
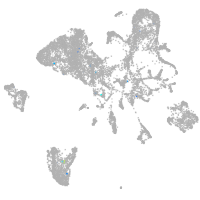
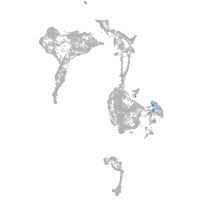
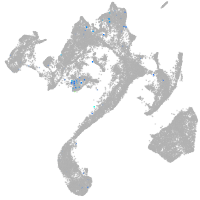
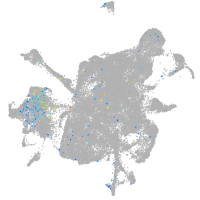
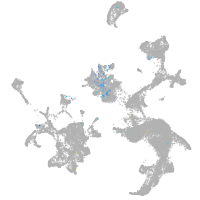
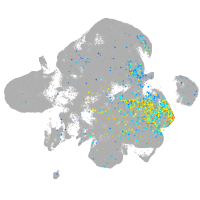
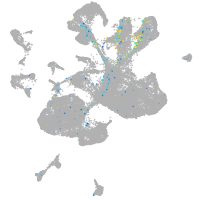
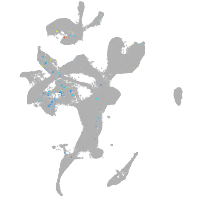
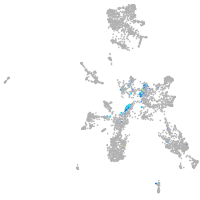
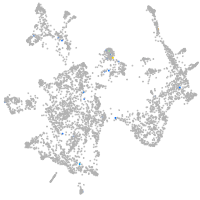
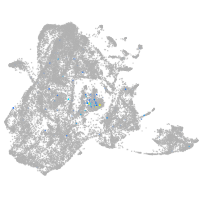
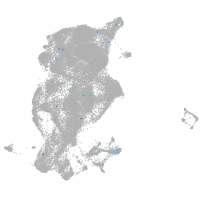
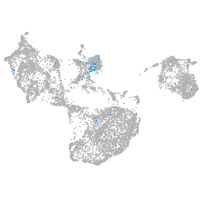
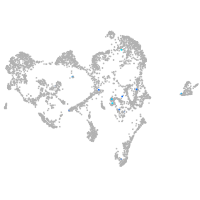
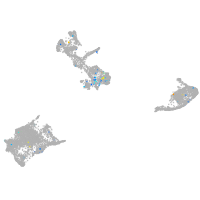
neuronal differentiation 6b
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| neurod6a | 0.384 | sparc | -0.059 |
| neurod2 | 0.331 | btg2 | -0.055 |
| neurod1 | 0.269 | eef1da | -0.047 |
| nfixb | 0.249 | rps17 | -0.045 |
| bhlhe22 | 0.227 | sdc4 | -0.045 |
| mllt11 | 0.225 | ier2b | -0.045 |
| eomesa | 0.213 | aldob | -0.044 |
| NPC1L1 | 0.212 | rpl23a | -0.044 |
| omga | 0.211 | rps6 | -0.044 |
| elavl4 | 0.210 | cebpb | -0.044 |
| cyp11a2 | 0.209 | prdx5 | -0.043 |
| si:ch211-233h19.2 | 0.206 | slc38a5b | -0.043 |
| zgc:153426 | 0.203 | rps7 | -0.042 |
| tuba1c | 0.203 | XLOC-044191 | -0.042 |
| XLOC-023835 | 0.201 | wu:fb18f06 | -0.042 |
| si:ch73-305o9.3 | 0.199 | dnase1l4.1 | -0.042 |
| dpysl3 | 0.194 | egfl6 | -0.042 |
| XLOC-008016 | 0.192 | tpt1 | -0.042 |
| cart1 | 0.190 | itgav | -0.041 |
| CR786576.1 | 0.190 | arhgef5 | -0.041 |
| ek1 | 0.188 | carhsp1 | -0.041 |
| wbp2 | 0.188 | rnaseka | -0.041 |
| stmn1b | 0.187 | col1a1a | -0.041 |
| camk2d2 | 0.185 | epcam | -0.041 |
| scrt1a | 0.184 | mcl1b | -0.041 |
| gng3 | 0.184 | aqp3a | -0.040 |
| evlb | 0.183 | cfl1l | -0.040 |
| mapk11 | 0.182 | zgc:158343 | -0.040 |
| cntnap5a | 0.182 | eno3 | -0.039 |
| stmn2a | 0.181 | rpl13 | -0.039 |
| gpm6ab | 0.180 | jun | -0.039 |
| dpysl2b | 0.180 | romo1 | -0.039 |
| pimr68 | 0.179 | cldni | -0.039 |
| elavl3 | 0.179 | ier2a | -0.038 |
| spock1 | 0.174 | pnp5a | -0.038 |