neuralized E3 ubiquitin protein ligase 1Ab
ZFIN 
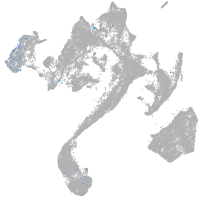
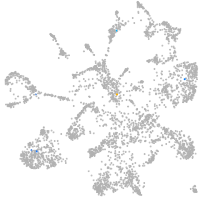
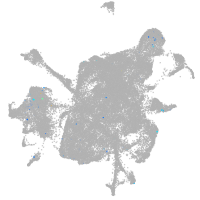
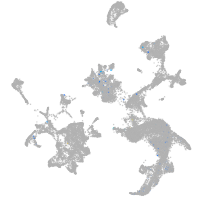
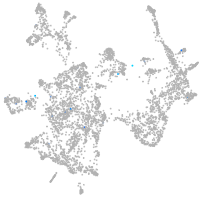
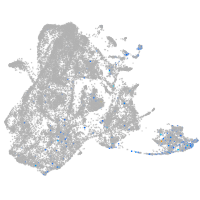
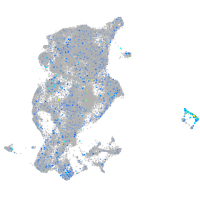
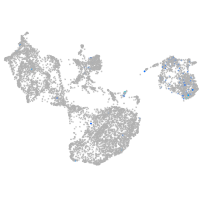
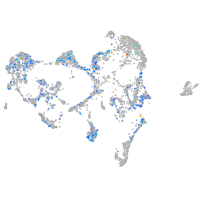
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tfap2e | 0.260 | hmgb2a | -0.096 |
| slc6a1a | 0.223 | fabp7a | -0.055 |
| tfap2c | 0.205 | ahcy | -0.052 |
| fosl2 | 0.187 | mdka | -0.049 |
| slc32a1 | 0.182 | msi1 | -0.047 |
| LOC101886114 | 0.180 | stmn1a | -0.047 |
| c1ql1a | 0.177 | pcna | -0.046 |
| tfap2a | 0.176 | msna | -0.046 |
| pax10 | 0.170 | nutf2l | -0.044 |
| syt1a | 0.169 | eef1da | -0.043 |
| XLOC-023761 | 0.169 | hmgb2b | -0.043 |
| meis2a | 0.168 | neurod4 | -0.042 |
| stxbp1a | 0.166 | dut | -0.042 |
| zmat4a | 0.165 | lbr | -0.042 |
| tfap2b | 0.165 | rrm1 | -0.041 |
| olfm2a | 0.162 | hes6 | -0.041 |
| celf5a | 0.159 | otx5 | -0.041 |
| znf385a | 0.159 | crx | -0.040 |
| mir181b-1 | 0.158 | mki67 | -0.040 |
| stx1b | 0.154 | ccnd1 | -0.040 |
| slc35g2b | 0.152 | syt5b | -0.040 |
| sv2a | 0.145 | COX7A2 (1 of many) | -0.040 |
| syn2b | 0.145 | chaf1a | -0.039 |
| gad2 | 0.143 | her15.1 | -0.039 |
| lrtm1 | 0.143 | banf1 | -0.037 |
| notum1b | 0.143 | rrm2 | -0.037 |
| mir181b-3 | 0.143 | ccna2 | -0.037 |
| dscama | 0.140 | mcm7 | -0.037 |
| ppp1r14ba | 0.140 | tuba8l | -0.037 |
| eno2 | 0.140 | selenoh | -0.037 |
| cabp1b | 0.140 | tspan7 | -0.037 |
| XLOC-020666 | 0.134 | dek | -0.036 |
| cplx3b | 0.133 | zgc:110425 | -0.036 |
| zeb2b | 0.133 | cks1b | -0.036 |
| meis2b | 0.132 | rpa3 | -0.035 |