NIMA-related kinase 11
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia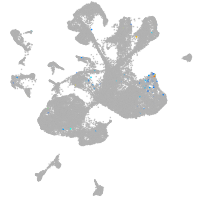 |
eye |
taste / olfactory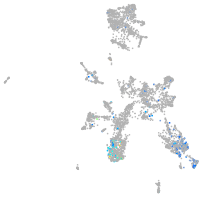 |
otic / lateral line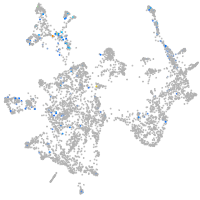 |
epidermis |
periderm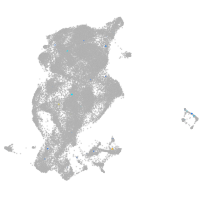 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cyp2k17 | 0.127 | gpm6aa | -0.025 |
| fam228a | 0.122 | gpm6ab | -0.022 |
| ompb | 0.122 | nova2 | -0.021 |
| got1l1 | 0.118 | marcksl1a | -0.021 |
| tekt2 | 0.117 | hbbe1.3 | -0.020 |
| capsla | 0.112 | hbae3 | -0.019 |
| vsig8a | 0.109 | hbae1.1 | -0.018 |
| ropn1l | 0.108 | celf2 | -0.017 |
| tekt1 | 0.108 | cspg5a | -0.017 |
| dzip1l | 0.105 | mdkb | -0.017 |
| lrrc23 | 0.105 | pvalb1 | -0.017 |
| mycbpap | 0.102 | hbbe1.1 | -0.016 |
| si:dkey-148f10.4 | 0.102 | pvalb2 | -0.016 |
| XLOC-024343 | 0.102 | fez1 | -0.015 |
| zgc:153142 | 0.102 | hbae1.3 | -0.015 |
| cnga4 | 0.102 | myt1b | -0.015 |
| cdc25d | 0.099 | ndrg4 | -0.015 |
| smkr1 | 0.098 | scrt2 | -0.015 |
| tekt4 | 0.098 | tubb5 | -0.015 |
| fabp10b | 0.097 | CU634008.1 | -0.015 |
| ppp1r42 | 0.097 | cadm3 | -0.014 |
| rsph14 | 0.097 | ckbb | -0.014 |
| capslb | 0.096 | hbbe1.2 | -0.014 |
| iqcg | 0.096 | hbbe2 | -0.014 |
| si:ch73-335l21.2 | 0.096 | krtt1c19e | -0.014 |
| enkur | 0.096 | snap25b | -0.014 |
| GBGT1 (1 of many) | 0.096 | stmn1b | -0.014 |
| spag6 | 0.095 | aep1 | -0.013 |
| CFAP77 | 0.094 | CU467822.1 | -0.013 |
| pih1d3 | 0.094 | cyt1l | -0.013 |
| spata18 | 0.092 | dbn1 | -0.013 |
| pacrg | 0.090 | elavl3 | -0.013 |
| emb | 0.088 | slc32a1 | -0.013 |
| lrrc51 | 0.088 | sox4a | -0.013 |
| ppil6 | 0.088 | tuba1c | -0.013 |




