NEDD4 E3 ubiquitin protein ligase a
ZFIN 
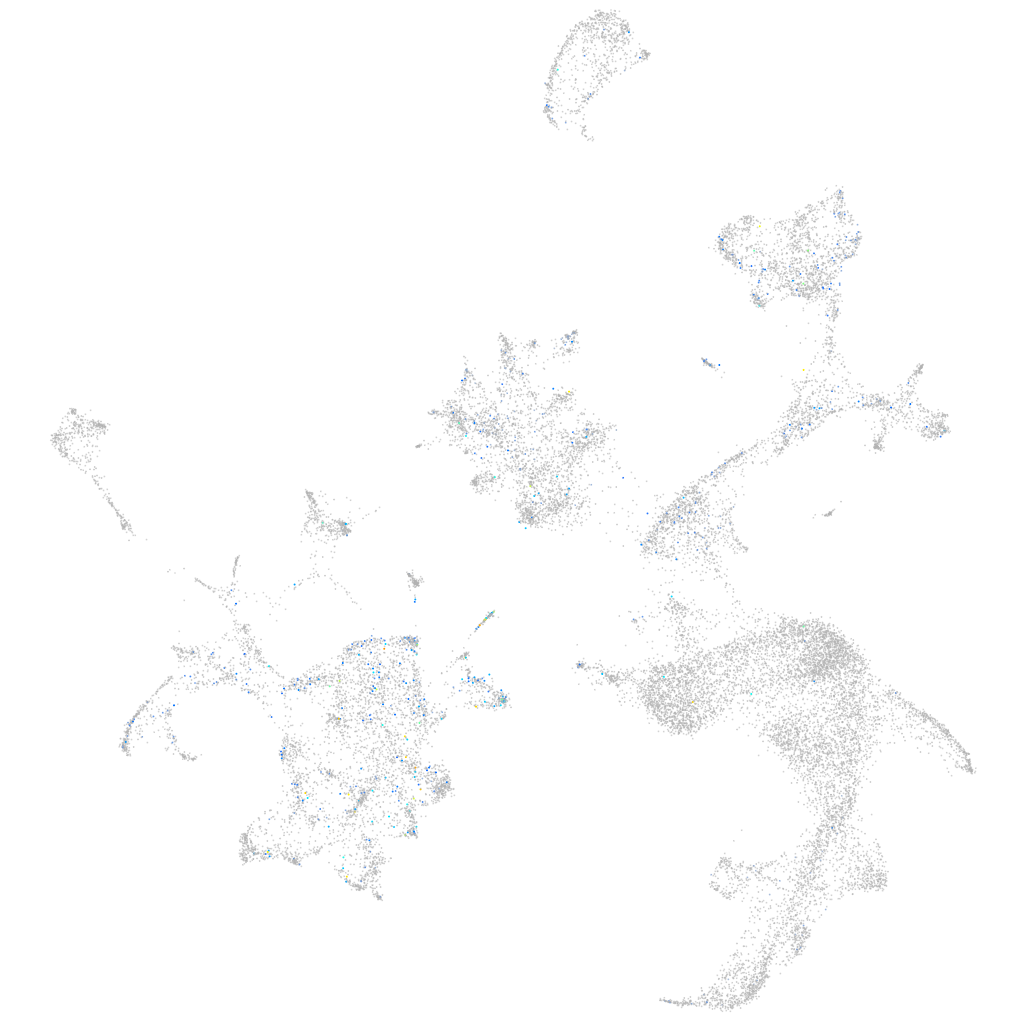
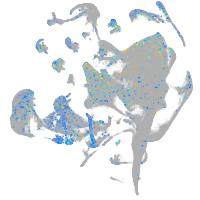
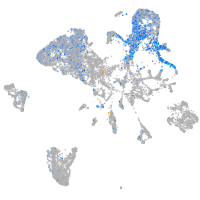
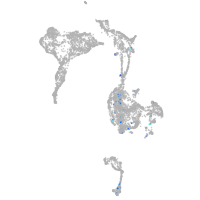
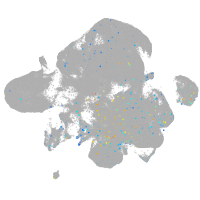
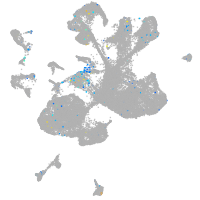
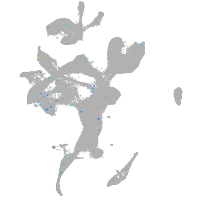
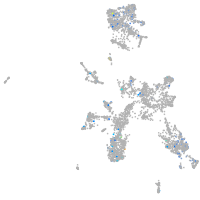
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| b3gnt7 | 0.124 | hbae1.3 | -0.087 |
| mrc1a | 0.114 | hbae1.1 | -0.087 |
| cpn1 | 0.111 | hbbe1.3 | -0.084 |
| cd81a | 0.110 | hbae3 | -0.082 |
| stab1 | 0.109 | hbbe2 | -0.081 |
| cd63 | 0.106 | cahz | -0.080 |
| calcrlb | 0.104 | hbbe1.1 | -0.080 |
| tgfbr2b | 0.104 | hemgn | -0.079 |
| marcksl1a | 0.104 | alas2 | -0.076 |
| ramp2 | 0.104 | blvrb | -0.075 |
| BX510949.1 | 0.103 | nt5c2l1 | -0.071 |
| myct1a | 0.102 | hbbe1.2 | -0.071 |
| hyal2b | 0.100 | fth1a | -0.070 |
| ecscr | 0.100 | prdx2 | -0.069 |
| ctsla | 0.099 | tspo | -0.068 |
| tpcn3 | 0.099 | creg1 | -0.065 |
| LOC101882910 | 0.098 | zgc:163057 | -0.064 |
| tpte | 0.098 | plac8l1 | -0.064 |
| stab2 | 0.098 | slc4a1a | -0.063 |
| CU984579.1 | 0.097 | si:ch211-207c6.2 | -0.062 |
| CR931782.2 | 0.097 | nmt1b | -0.061 |
| vat1 | 0.097 | si:ch211-250g4.3 | -0.060 |
| LO017955.1 | 0.096 | zgc:56095 | -0.058 |
| lgmn | 0.096 | tmod4 | -0.057 |
| ednraa | 0.096 | epb41b | -0.053 |
| lyve1b | 0.095 | hbae5 | -0.050 |
| LOC110437899 | 0.094 | gpx1a | -0.048 |
| CU915818.1 | 0.094 | hbbe3 | -0.048 |
| lamp2 | 0.093 | add2 | -0.048 |
| LOC108179280 | 0.092 | ucp3 | -0.044 |
| dusp5 | 0.091 | rfesd | -0.044 |
| flt4 | 0.091 | klf1 | -0.043 |
| rab8b | 0.091 | CR293511.1 | -0.043 |
| zgc:110239 | 0.091 | sptb | -0.042 |
| fstl1b | 0.091 | rhd | -0.040 |