nectin cell adhesion molecule 3b
ZFIN 
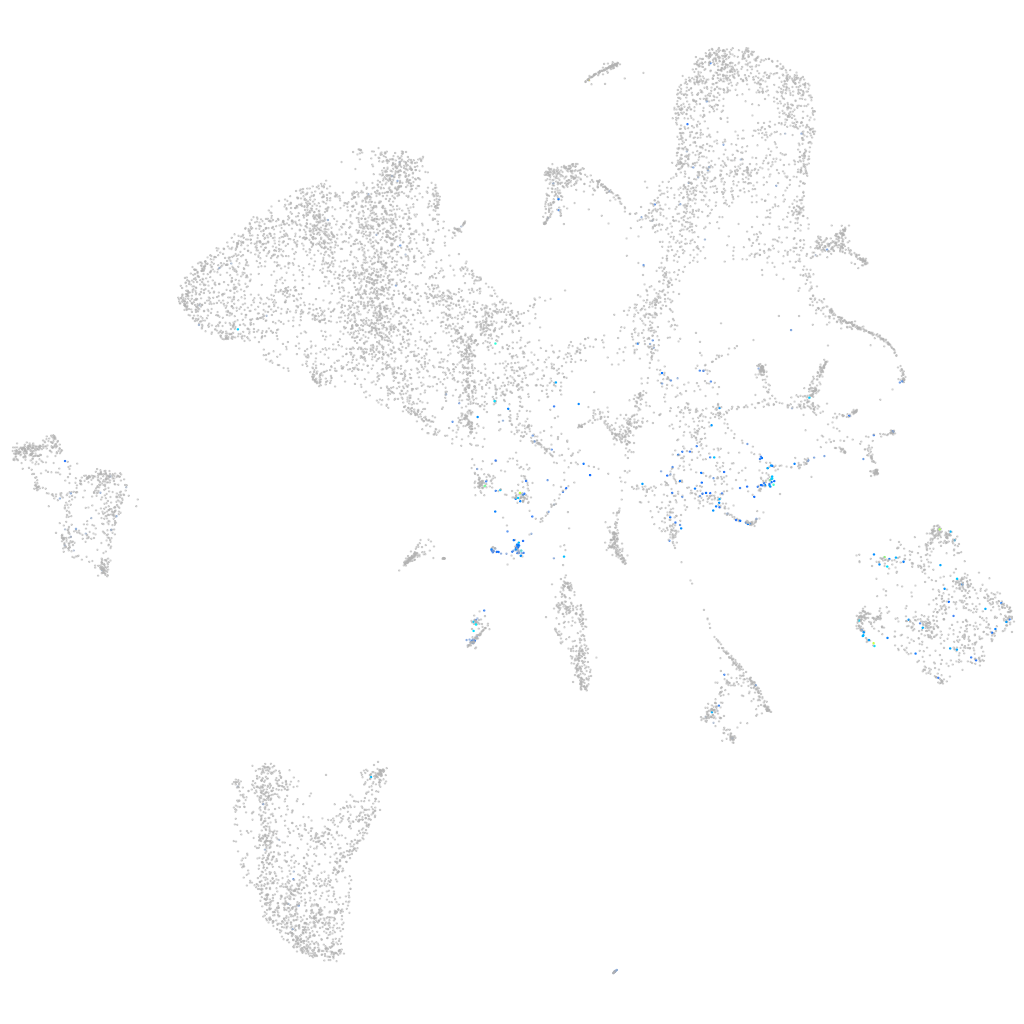
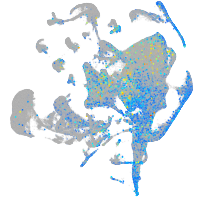
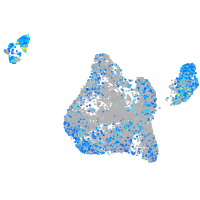
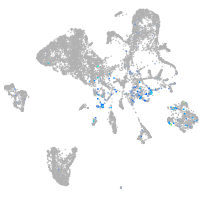
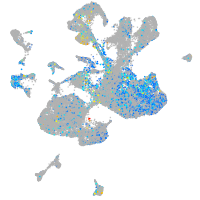
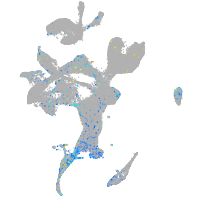
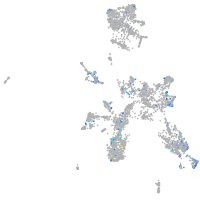
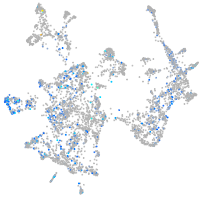
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gdf6b | 0.236 | ahcy | -0.131 |
| ndnf | 0.204 | gapdh | -0.128 |
| tpm4a | 0.202 | gamt | -0.122 |
| ptgs1 | 0.194 | sod1 | -0.117 |
| col8a1a | 0.190 | aldob | -0.114 |
| vcana | 0.190 | fbp1b | -0.111 |
| fgfrl1b | 0.184 | dap | -0.111 |
| hoxc6b | 0.180 | glud1b | -0.111 |
| cx43.4 | 0.176 | gpx4a | -0.110 |
| apela | 0.174 | gatm | -0.109 |
| qkia | 0.172 | nupr1b | -0.108 |
| hapln1b | 0.170 | mat1a | -0.105 |
| bcam | 0.170 | gstt1a | -0.104 |
| prxl2b | 0.168 | apoa4b.1 | -0.102 |
| CABZ01075068.1 | 0.166 | scp2a | -0.102 |
| plod2 | 0.165 | apoc2 | -0.100 |
| pcdh19 | 0.165 | apoa1b | -0.098 |
| her12 | 0.164 | tpi1b | -0.098 |
| hhip | 0.164 | pgk1 | -0.097 |
| si:ch211-222l21.1 | 0.162 | eno3 | -0.097 |
| id3 | 0.160 | atp5if1b | -0.097 |
| sept15 | 0.160 | suclg1 | -0.097 |
| lsp1 | 0.160 | gstp1 | -0.095 |
| angptl6 | 0.157 | mdh1aa | -0.095 |
| nkx6.2 | 0.157 | sdr16c5b | -0.093 |
| fbn2b | 0.156 | pgm1 | -0.092 |
| ntd5 | 0.156 | suclg2 | -0.092 |
| BX001014.2 | 0.156 | gnmt | -0.090 |
| hmgb3a | 0.156 | rdh1 | -0.090 |
| crtap | 0.155 | g6pca.2 | -0.090 |
| hp1bp3 | 0.155 | afp4 | -0.090 |
| LOC103909694 | 0.152 | tdo2a | -0.090 |
| pltp | 0.152 | agxtb | -0.089 |
| sall4 | 0.152 | bhmt | -0.089 |
| fstl1b | 0.151 | msrb2 | -0.089 |