nectin cell adhesion molecule 1a
ZFIN 
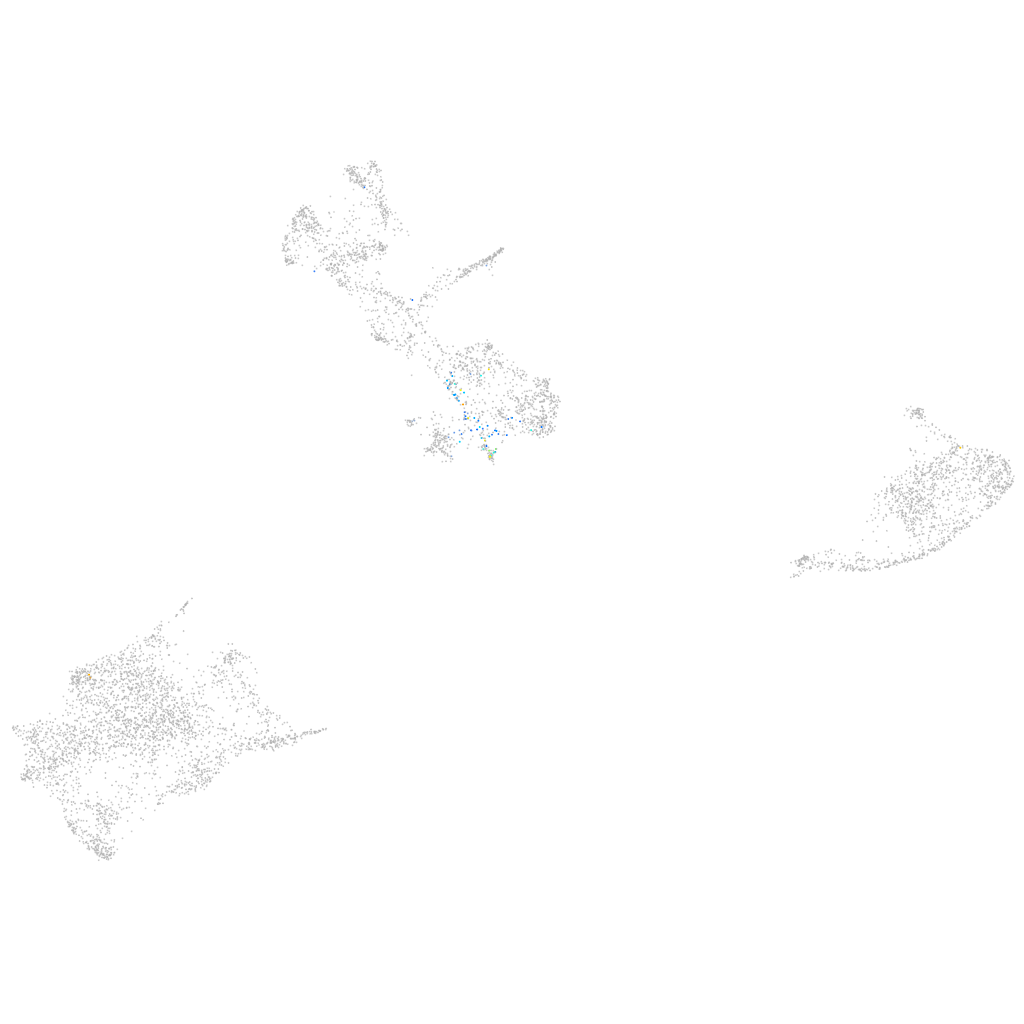
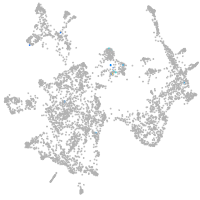
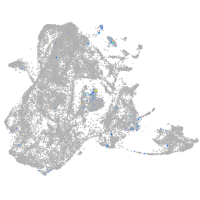
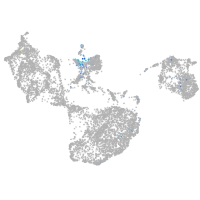
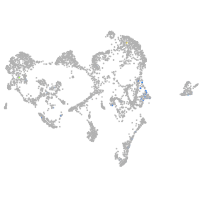
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| syt5b | 0.333 | tspan36 | -0.070 |
| otx5 | 0.309 | gstp1 | -0.066 |
| si:ch73-256g18.2 | 0.279 | syngr1a | -0.063 |
| rs1a | 0.273 | gpr143 | -0.059 |
| myh11b | 0.236 | pcbd1 | -0.058 |
| crx | 0.233 | cst14a.2 | -0.058 |
| vsx1 | 0.232 | rab38 | -0.055 |
| lrit1b | 0.230 | rab32a | -0.054 |
| gnb3a | 0.222 | akr1b1 | -0.054 |
| tmem151bb | 0.222 | zgc:110239 | -0.054 |
| snap25b | 0.219 | ctsd | -0.053 |
| nrn1lb | 0.218 | actb2 | -0.053 |
| scg3 | 0.218 | bace2 | -0.052 |
| cabp2a | 0.218 | paics | -0.051 |
| foxg1b | 0.214 | tmem243a | -0.051 |
| arl3l1 | 0.209 | rnaseka | -0.050 |
| pimr178 | 0.208 | qdpra | -0.050 |
| calb2b | 0.206 | eno3 | -0.050 |
| cplx4a | 0.205 | zgc:165573 | -0.050 |
| UNC13B | 0.203 | prdx6 | -0.049 |
| guca1g | 0.203 | gart | -0.049 |
| bhlhe23 | 0.203 | ctsla | -0.049 |
| samsn1a | 0.201 | slc2a15a | -0.047 |
| CABZ01041495.1 | 0.200 | ucp2 | -0.047 |
| ppm1na | 0.198 | slc45a2 | -0.047 |
| cnga3b | 0.198 | prdx1 | -0.046 |
| atp2b1b | 0.197 | slc2a11b | -0.046 |
| RIC3 | 0.197 | agtrap | -0.046 |
| gabrr3a | 0.197 | LOC103910009 | -0.045 |
| cadm3 | 0.196 | mitfa | -0.045 |
| arhgef25b | 0.195 | slc3a2a | -0.045 |
| CABZ01038709.1 | 0.194 | si:dkey-21a6.5 | -0.044 |
| anp32e | 0.194 | tmsb4x | -0.044 |
| neurod4 | 0.191 | CABZ01048402.2 | -0.043 |
| taar10d | 0.191 | slc38a5b | -0.043 |