NECAP endocytosis associated 1
ZFIN 
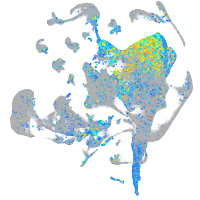
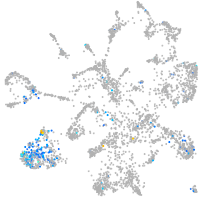
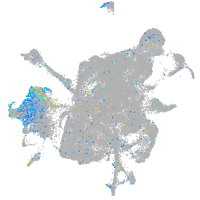
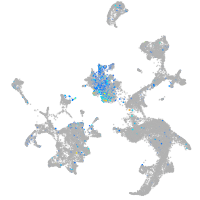
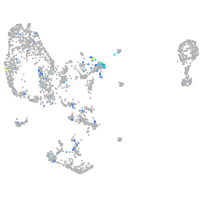
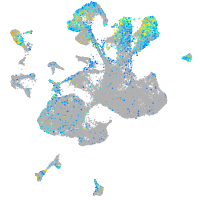
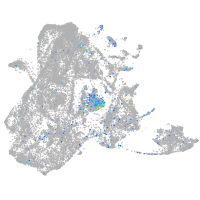
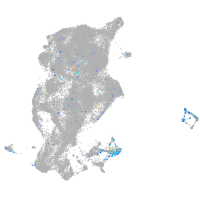
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.396 | gapdh | -0.195 |
| rnasekb | 0.342 | ahcy | -0.190 |
| pcsk1nl | 0.335 | gamt | -0.189 |
| gapdhs | 0.326 | gatm | -0.180 |
| scgn | 0.324 | fbp1b | -0.167 |
| si:dkey-153k10.9 | 0.324 | mat1a | -0.160 |
| neurod1 | 0.323 | apoa4b.1 | -0.156 |
| pax6b | 0.317 | scp2a | -0.155 |
| vat1 | 0.310 | gstr | -0.155 |
| syt1a | 0.308 | aldob | -0.155 |
| mir7a-1 | 0.305 | aldh6a1 | -0.153 |
| gnao1a | 0.303 | aqp12 | -0.152 |
| calm1b | 0.296 | bhmt | -0.152 |
| vamp2 | 0.296 | gstt1a | -0.151 |
| cpe | 0.285 | apoc2 | -0.151 |
| atpv0e2 | 0.283 | cx32.3 | -0.151 |
| pcsk1 | 0.282 | aldh7a1 | -0.149 |
| zgc:101731 | 0.281 | apoa1b | -0.145 |
| lysmd2 | 0.278 | afp4 | -0.143 |
| cplx2l | 0.276 | agxtb | -0.142 |
| egr4 | 0.275 | pnp4b | -0.141 |
| insm1a | 0.275 | glud1b | -0.138 |
| snap25a | 0.274 | suclg2 | -0.137 |
| rprmb | 0.274 | abat | -0.136 |
| gpc1a | 0.273 | gcshb | -0.135 |
| aldocb | 0.273 | si:dkey-16p21.8 | -0.135 |
| id4 | 0.272 | nipsnap3a | -0.134 |
| atp6v0cb | 0.272 | slc27a2a | -0.134 |
| tspan7b | 0.270 | agxta | -0.133 |
| scg2a | 0.270 | phyhd1 | -0.133 |
| hsbp1a | 0.270 | nupr1b | -0.132 |
| c2cd4a | 0.267 | g6pca.2 | -0.132 |
| pcsk2 | 0.267 | apobb.1 | -0.132 |
| isl1 | 0.259 | sult2st2 | -0.131 |
| nkx2.2a | 0.259 | mgst1.2 | -0.131 |