nanos homolog 1
ZFIN 
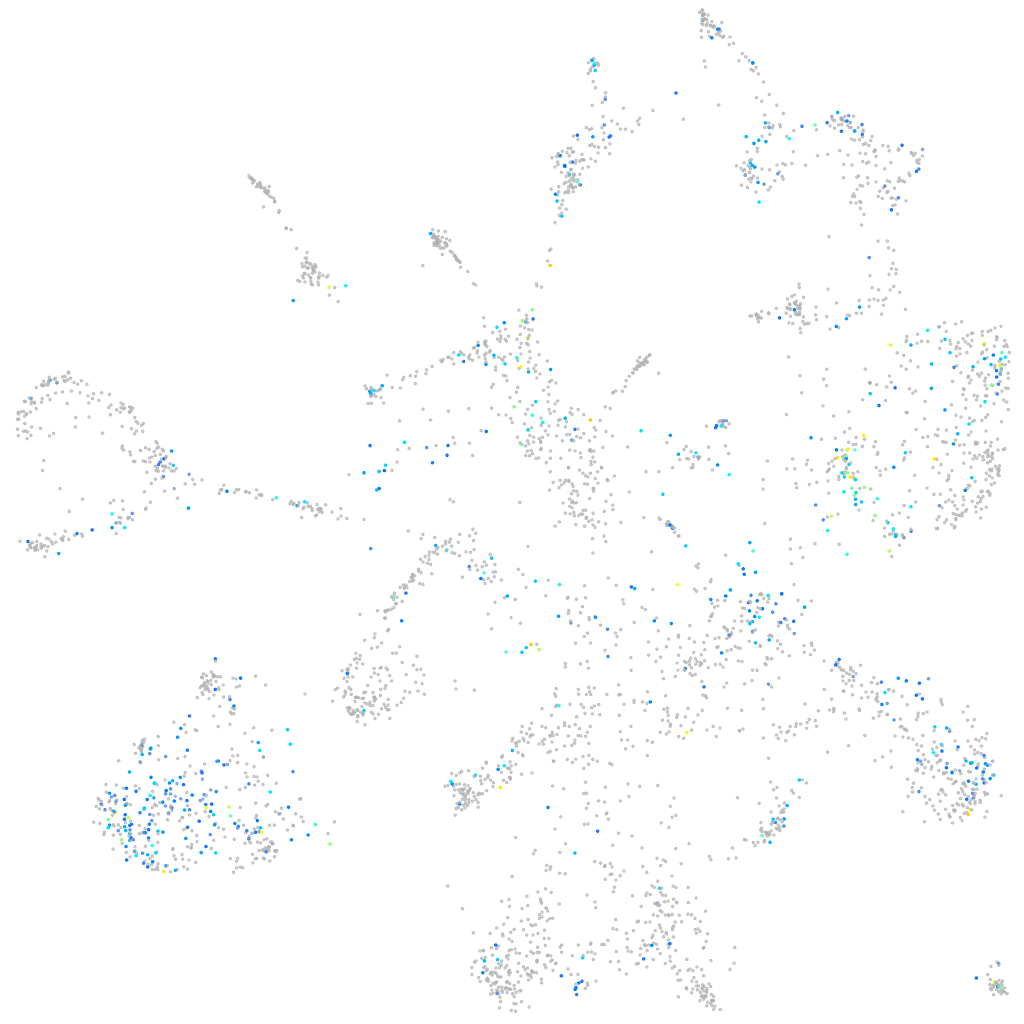
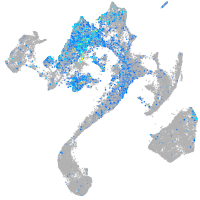
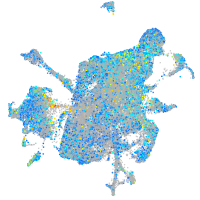
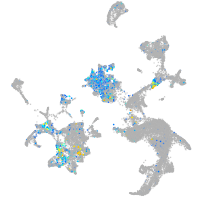
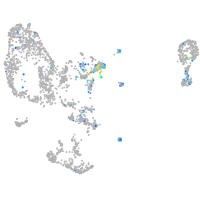
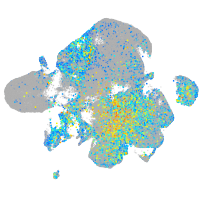
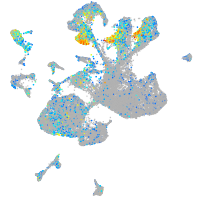
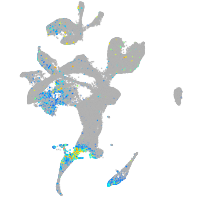
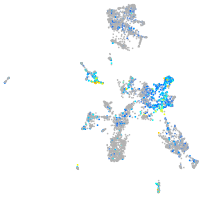
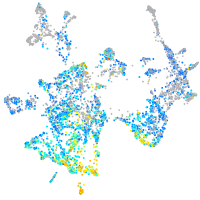
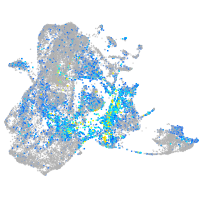
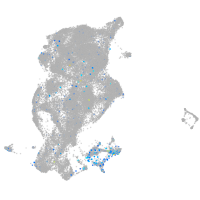
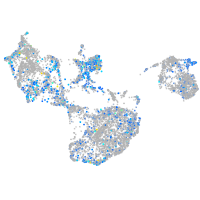
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nova2 | 0.176 | col6a2 | -0.113 |
| stmn1b | 0.170 | aoc2 | -0.103 |
| rtn1b | 0.159 | si:ch211-62a1.3 | -0.100 |
| elavl4 | 0.155 | BX088707.3 | -0.099 |
| si:ch211-137a8.4 | 0.154 | col6a1 | -0.093 |
| hunk | 0.153 | tpm1 | -0.091 |
| elavl3 | 0.152 | ak1 | -0.087 |
| tmsb | 0.151 | myl9a | -0.085 |
| sox4a | 0.150 | igfbp7 | -0.084 |
| scn8aa | 0.143 | rbpms2a | -0.084 |
| syt1a | 0.142 | foxf1 | -0.083 |
| stmn2a | 0.142 | rgs5a | -0.083 |
| gpm6ab | 0.141 | myl9b | -0.083 |
| gng3 | 0.140 | mylkb | -0.080 |
| rbfox1 | 0.140 | mamdc2a | -0.079 |
| tuba1c | 0.139 | myh11a | -0.079 |
| pax2a | 0.137 | ntn5 | -0.078 |
| rtn1a | 0.136 | nid1b | -0.075 |
| gabrb4 | 0.136 | tagln | -0.074 |
| snap25a | 0.135 | lama4 | -0.073 |
| pik3r3b | 0.134 | bgnb | -0.072 |
| myt1la | 0.133 | myl6 | -0.072 |
| kcnn1a | 0.132 | itga1 | -0.070 |
| epb41a | 0.131 | cnn1b | -0.070 |
| brinp2 | 0.131 | itga4 | -0.070 |
| zgc:65894 | 0.130 | acta2 | -0.069 |
| adgrb1a | 0.129 | desmb | -0.069 |
| stxbp1a | 0.129 | nkx2.3 | -0.069 |
| vamp2 | 0.128 | parvab | -0.069 |
| tmeff1b | 0.128 | smtna | -0.068 |
| sncb | 0.128 | col4a1 | -0.067 |
| CABZ01061349.1 | 0.126 | slc2a1b | -0.067 |
| FO082781.1 | 0.126 | pdgfrb | -0.066 |
| gpm6aa | 0.124 | lamc3 | -0.066 |
| fthl27 | 0.123 | gucy1a1 | -0.065 |