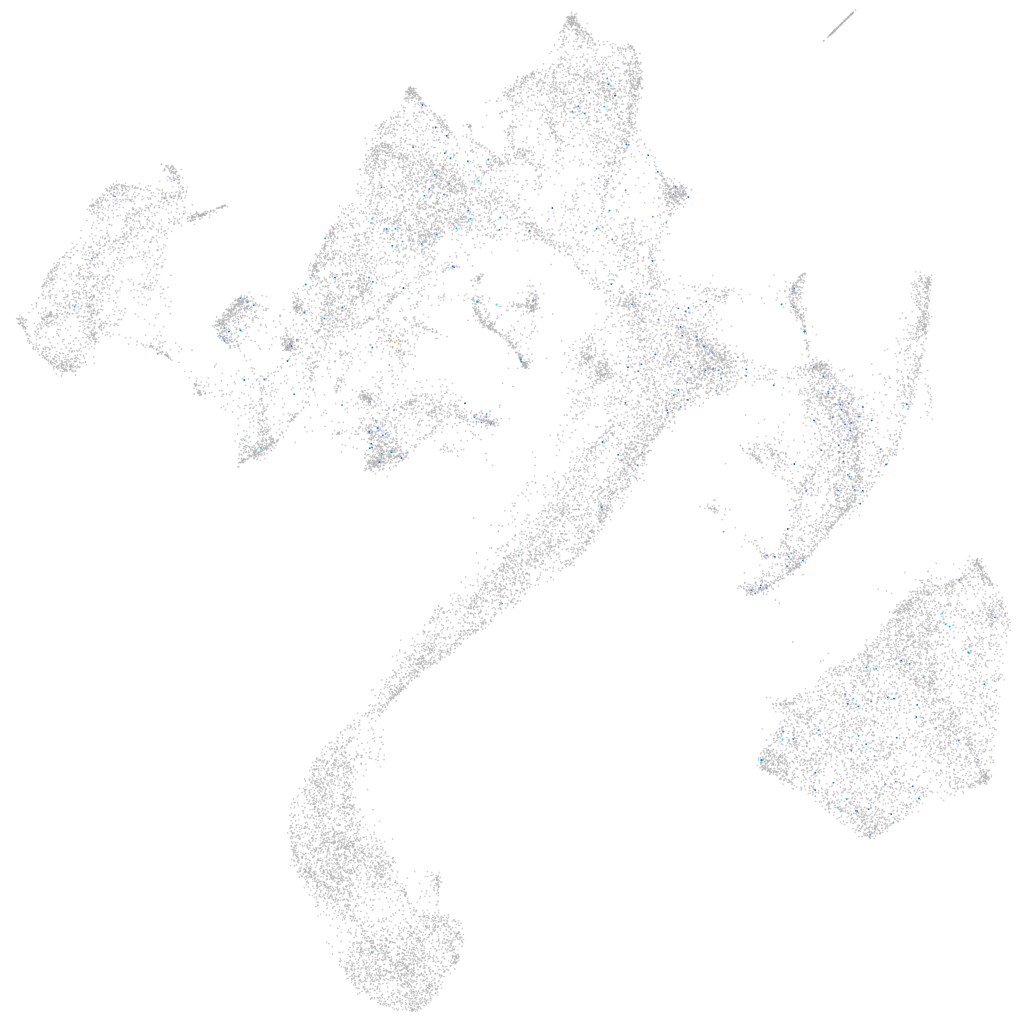
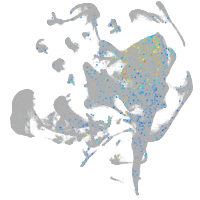
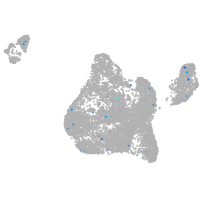
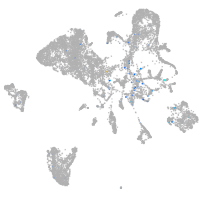
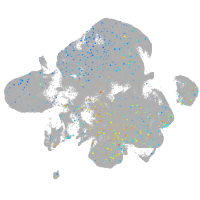
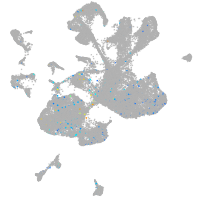
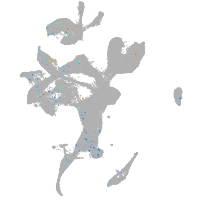
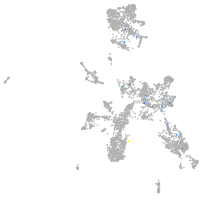
"nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b"
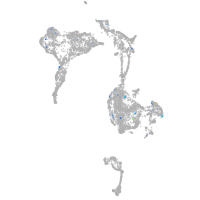
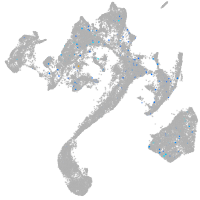
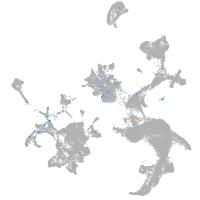
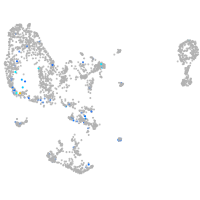
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpr153 | 0.140 | aldoab | -0.046 |
| LOC108191351 | 0.115 | actc1b | -0.045 |
| LOC101886244 | 0.108 | acta1b | -0.044 |
| ros1 | 0.096 | mybphb | -0.044 |
| dip2bb | 0.090 | ak1 | -0.043 |
| LOC101885655 | 0.089 | ckmb | -0.043 |
| CR626886.1 | 0.084 | ldb3a | -0.042 |
| CABZ01086139.1 | 0.083 | CABZ01078594.1 | -0.042 |
| CU207343.1 | 0.083 | atp2a1 | -0.041 |
| AL929396.1 | 0.082 | tnnc2 | -0.041 |
| LOC101885363 | 0.082 | gapdh | -0.041 |
| si:ch211-67f13.7 | 0.079 | ckma | -0.041 |
| LOC103909163 | 0.076 | ldb3b | -0.041 |
| or131-2 | 0.076 | tpma | -0.041 |
| spire2 | 0.075 | tmem38a | -0.041 |
| si:ch73-106l15.4 | 0.074 | srl | -0.040 |
| elmod1 | 0.074 | eno3 | -0.040 |
| BX004785.1 | 0.073 | ttn.2 | -0.039 |
| or119-2 | 0.071 | neb | -0.039 |
| gria2b | 0.071 | pabpc4 | -0.038 |
| zgc:174180 | 0.071 | hhatla | -0.038 |
| XLOC-013628 | 0.069 | mylpfa | -0.038 |
| foxg1d | 0.069 | klhl31 | -0.037 |
| RYR2 | 0.068 | gamt | -0.037 |
| lrrc4c | 0.067 | smyd1a | -0.037 |
| FP016247.1 | 0.067 | ttn.1 | -0.037 |
| shank2 | 0.066 | actn3b | -0.037 |
| LOC110440219 | 0.065 | ank1a | -0.037 |
| cnih2 | 0.065 | rtn2a | -0.037 |
| FP016005.1 | 0.065 | myl1 | -0.037 |
| GRIK2 | 0.064 | zgc:101853 | -0.036 |
| vtnb | 0.063 | CABZ01072309.1 | -0.036 |
| si:ch211-26b3.4 | 0.061 | cav3 | -0.036 |
| BX324134.1 | 0.061 | desma | -0.036 |
| syt4 | 0.060 | sh3bgr | -0.036 |