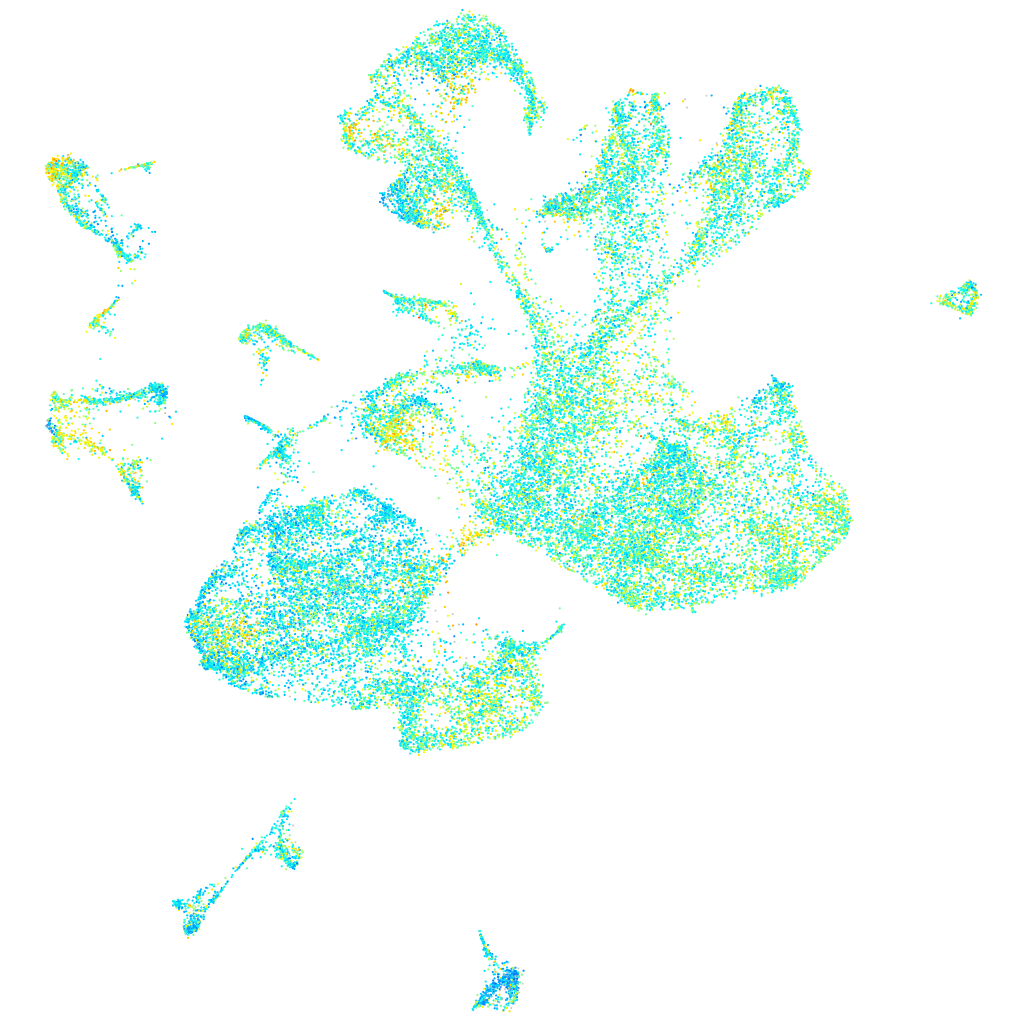
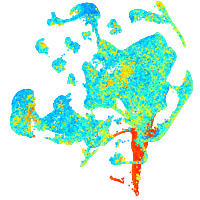
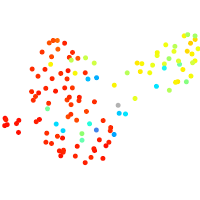
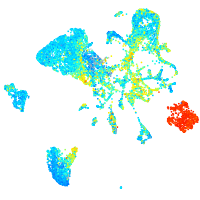
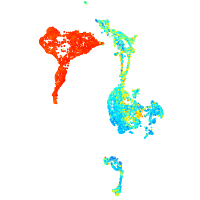
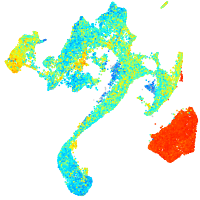
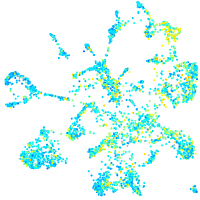
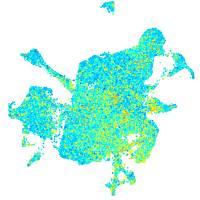
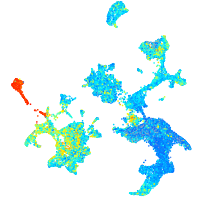
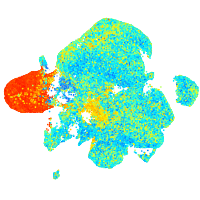
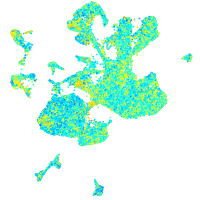
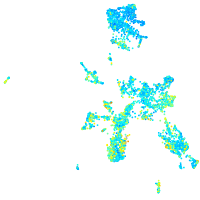
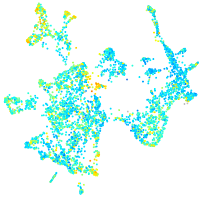
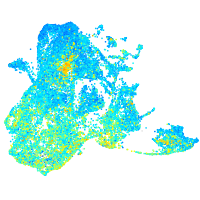
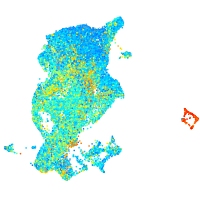
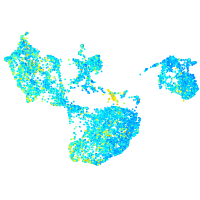
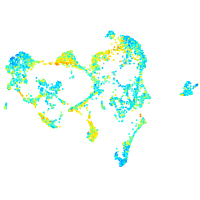
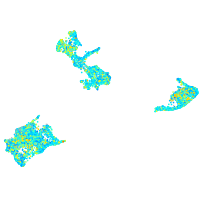
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| NC-002333.17 | 0.439 | pvalb1 | -0.201 |
| mt-co2 | 0.414 | pvalb2 | -0.187 |
| mt-co1 | 0.394 | fabp7a | -0.180 |
| COX3 | 0.392 | apoa2 | -0.175 |
| mt-cyb | 0.352 | CU467822.1 | -0.172 |
| CABZ01072614.1 | 0.262 | zgc:165604 | -0.169 |
| CABZ01075068.1 | 0.210 | rlbp1a | -0.167 |
| mt-nd3 | 0.197 | ca14 | -0.165 |
| mt-nd2 | 0.196 | apoa1b | -0.164 |
| rpl3 | 0.195 | glula | -0.163 |
| mt-nd4 | 0.192 | hyal6 | -0.154 |
| mt-atp6 | 0.183 | si:dkey-16p21.8 | -0.153 |
| mt-nd1 | 0.174 | actc1b | -0.151 |
| ppiaa | 0.161 | rhbg | -0.149 |
| LOC110439372 | 0.144 | cavin2a | -0.147 |
| zgc:158463 | 0.144 | dap1b | -0.145 |
| rpl4 | 0.141 | cebpd | -0.142 |
| oc90 | 0.138 | cdo1 | -0.141 |
| rps18 | 0.136 | efhd1 | -0.140 |
| mt-nd5 | 0.136 | acbd7 | -0.140 |
| igf2bp1 | 0.131 | cahz | -0.137 |
| cdh2 | 0.130 | crabp1a | -0.136 |
| ankrd11 | 0.129 | prss59.2 | -0.135 |
| hsp90ab1 | 0.128 | ckbb | -0.133 |
| trim71 | 0.127 | ppdpfb | -0.132 |
| zbtb16a | 0.126 | rbpms2b | -0.131 |
| hoxb1b | 0.125 | slc2a1a | -0.131 |
| tuba1a | 0.125 | syngr2b | -0.130 |
| greb1l | 0.124 | fgf24 | -0.127 |
| acin1a | 0.122 | mt2 | -0.125 |
| lin28a | 0.120 | LOC101886429 | -0.124 |
| hoxb3a | 0.120 | cldn5b | -0.123 |
| rps6 | 0.117 | CELA1 (1 of many) | -0.122 |
| AL935186.9 | 0.116 | slmapb | -0.121 |
| rpl15 | 0.116 | sod1 | -0.120 |