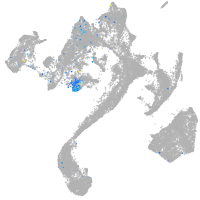
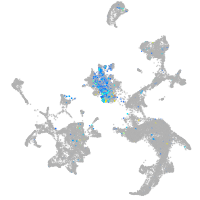
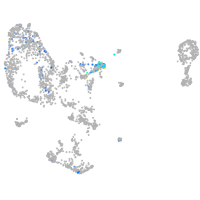
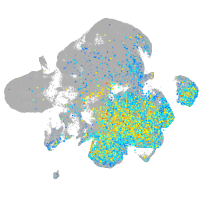
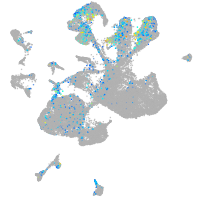
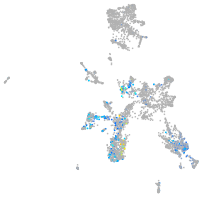
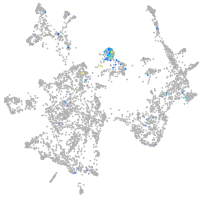
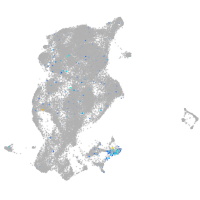
"myelin transcription factor 1-like, a"
ZFIN 
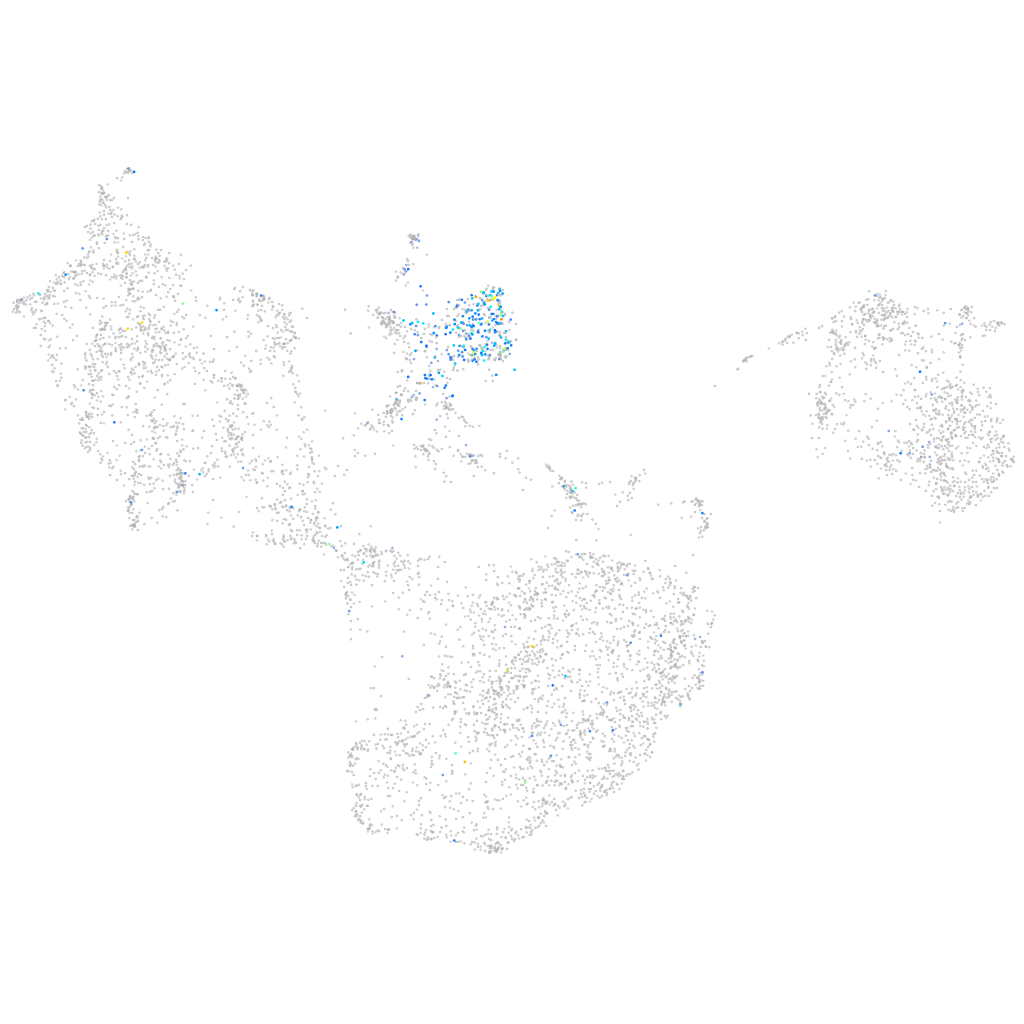
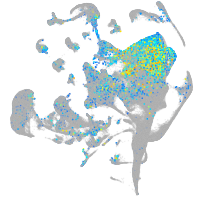
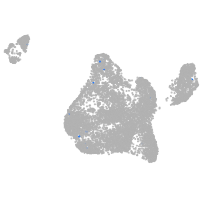
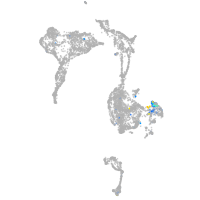
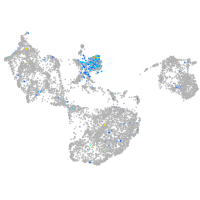
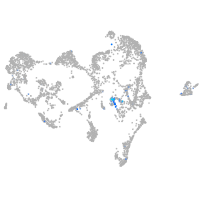
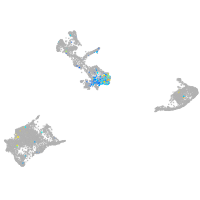
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stmn1b | 0.473 | sparc | -0.135 |
| tuba1c | 0.452 | s100a10b | -0.125 |
| rtn1a | 0.450 | zgc:153867 | -0.124 |
| rtn1b | 0.450 | anxa1a | -0.120 |
| gpm6aa | 0.444 | krt4 | -0.120 |
| stxbp1a | 0.432 | epcam | -0.119 |
| sncb | 0.431 | cfl1l | -0.117 |
| stx1b | 0.429 | pfn1 | -0.116 |
| zgc:153426 | 0.425 | actb2 | -0.115 |
| nova2 | 0.424 | tmsb1 | -0.113 |
| cspg5a | 0.424 | cldn1 | -0.113 |
| gng3 | 0.420 | cldni | -0.112 |
| elavl3 | 0.418 | lgals3b | -0.111 |
| stmn2a | 0.416 | cyt1 | -0.110 |
| gpm6ab | 0.410 | krt8 | -0.107 |
| ywhag2 | 0.410 | tuba8l2 | -0.107 |
| maptb | 0.405 | ahnak | -0.107 |
| elavl4 | 0.404 | pleca | -0.106 |
| atp6v0cb | 0.402 | ccng1 | -0.105 |
| tubb5 | 0.394 | hdlbpa | -0.104 |
| fez1 | 0.386 | ecrg4b | -0.103 |
| gap43 | 0.382 | GCA | -0.102 |
| vamp2 | 0.381 | pak2a | -0.102 |
| tmsb2 | 0.378 | icn | -0.102 |
| cdk5r1b | 0.377 | hbegfa | -0.101 |
| mllt11 | 0.377 | pycard | -0.100 |
| si:dkey-276j7.1 | 0.375 | eno3 | -0.100 |
| gnao1a | 0.373 | spaca4l | -0.099 |
| sv2a | 0.366 | zgc:153284 | -0.099 |
| aplp1 | 0.364 | dnase1l4.1 | -0.099 |
| nlgn2b | 0.364 | eef1a1l1 | -0.098 |
| ppp1r14ba | 0.360 | cd9b | -0.098 |
| snap25a | 0.358 | anxa2a | -0.098 |
| map1aa | 0.357 | tagln2 | -0.098 |
| zgc:65894 | 0.356 | zgc:162730 | -0.098 |