v-myb avian myeloblastosis viral oncogene homolog-like 1
ZFIN 
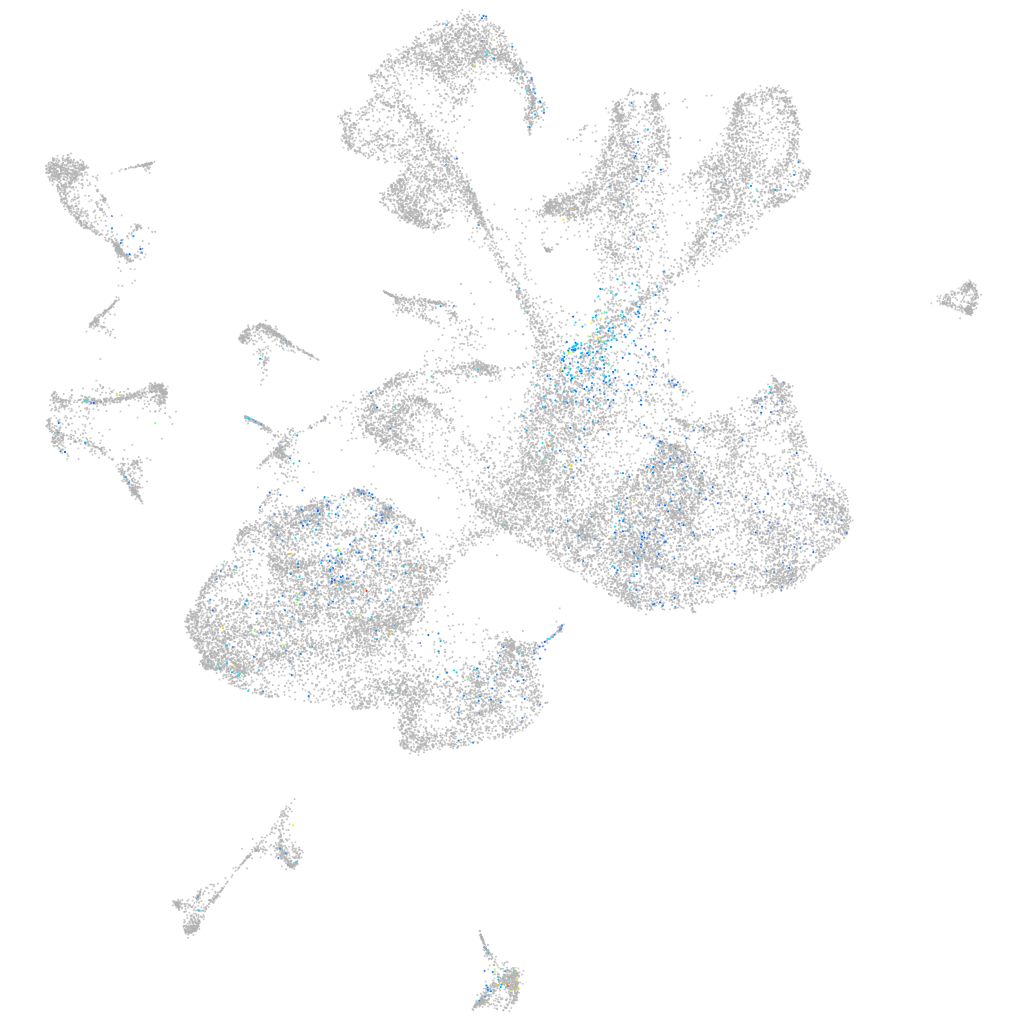
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-152f23.5 | 0.172 | marcksl1b | -0.047 |
| im:7152348 | 0.109 | snap25a | -0.044 |
| dlb | 0.109 | stmn2a | -0.044 |
| BX530077.1 | 0.100 | elavl4 | -0.044 |
| zgc:110540 | 0.099 | calm1a | -0.043 |
| dla | 0.098 | ywhag2 | -0.041 |
| pcdh18b | 0.097 | stx1b | -0.041 |
| tk1 | 0.097 | sncb | -0.041 |
| neurod4 | 0.096 | gng3 | -0.041 |
| insm1a | 0.094 | cplx2l | -0.041 |
| rrm2 | 0.091 | id4 | -0.040 |
| gadd45gb.1 | 0.091 | cplx2 | -0.039 |
| dck | 0.089 | map1aa | -0.039 |
| CABZ01005379.1 | 0.088 | calm2a | -0.038 |
| rrm1 | 0.088 | stxbp1a | -0.037 |
| her13 | 0.088 | zgc:65894 | -0.036 |
| ebf2 | 0.087 | sncgb | -0.036 |
| acap3b | 0.087 | eno2 | -0.035 |
| tyms | 0.085 | rtn1b | -0.035 |
| atoh1b | 0.085 | gap43 | -0.035 |
| insm1b | 0.084 | gpm6ab | -0.034 |
| prox1a | 0.083 | tuba2 | -0.034 |
| esco2 | 0.082 | slc18a3a | -0.034 |
| mibp | 0.082 | gabrb4 | -0.034 |
| notch1a | 0.082 | aplp1 | -0.034 |
| srrm4 | 0.081 | syt2a | -0.033 |
| nhlh2 | 0.081 | si:dkeyp-117h8.2 | -0.033 |
| inavaa | 0.081 | rbfox1 | -0.033 |
| dlc | 0.081 | tmem59l | -0.033 |
| slbp | 0.080 | ribc1 | -0.032 |
| znf367 | 0.079 | islr2 | -0.032 |
| pcna | 0.079 | fez1 | -0.032 |
| ccne2 | 0.078 | palm1a | -0.032 |
| fstl1a | 0.077 | syn2a | -0.031 |
| hes2.2 | 0.076 | cd81a | -0.031 |