mevalonate kinase
ZFIN 
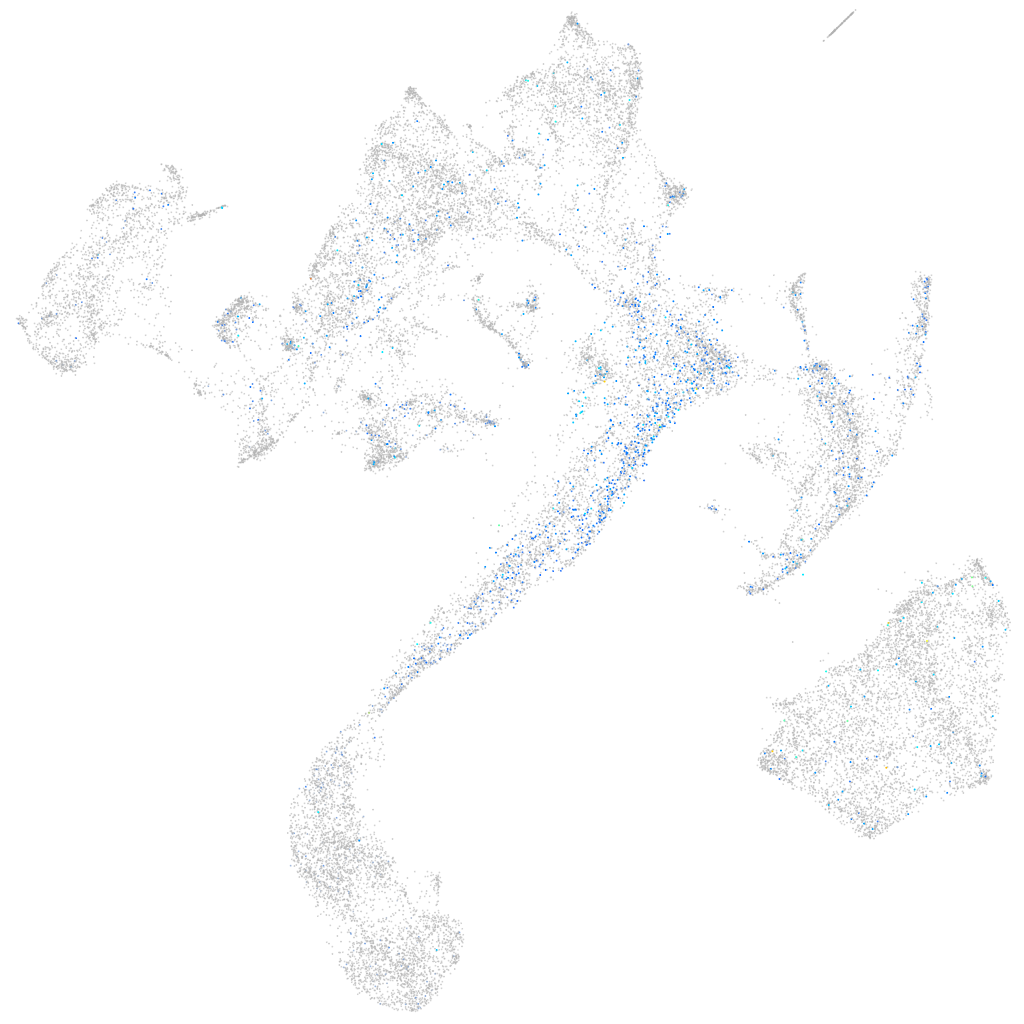
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mafbb | 0.145 | si:dkey-16p21.8 | -0.083 |
| bysl | 0.125 | pvalb1 | -0.072 |
| snu13b | 0.124 | actn3a | -0.069 |
| cct3 | 0.123 | bhmt | -0.069 |
| cct5 | 0.122 | pkmb | -0.068 |
| btf3l4 | 0.122 | pvalb2 | -0.068 |
| zbtb18 | 0.121 | eno3 | -0.068 |
| rbm24a | 0.121 | prx | -0.067 |
| cnbpa | 0.121 | tnnt3b | -0.067 |
| sirt7 | 0.120 | smyd1a | -0.067 |
| rpl7l1 | 0.120 | si:ch211-266g18.10 | -0.067 |
| eif4ebp1 | 0.119 | si:ch211-255p10.3 | -0.067 |
| tma16 | 0.119 | XLOC-025819 | -0.066 |
| cct7 | 0.119 | neb | -0.065 |
| tomm20a | 0.119 | si:ch73-367p23.2 | -0.065 |
| eif6 | 0.119 | casq1b | -0.065 |
| ptk2ab | 0.118 | myom1a | -0.064 |
| ftsj3 | 0.118 | ldb3b | -0.064 |
| emg1 | 0.117 | XLOC-005350 | -0.064 |
| nhp2 | 0.117 | tmod4 | -0.064 |
| pfdn6 | 0.116 | XLOC-006515 | -0.064 |
| hnrnpa0l | 0.115 | pgam2 | -0.064 |
| c1qbp | 0.114 | myom1b | -0.063 |
| pbdc1 | 0.114 | dhrs7cb | -0.063 |
| nifk | 0.113 | ckma | -0.063 |
| npm3 | 0.113 | XLOC-001975 | -0.062 |
| fam107b | 0.113 | glud1b | -0.062 |
| jam2a | 0.112 | ckmb | -0.061 |
| ssb | 0.112 | ndrg2 | -0.061 |
| rplp0 | 0.112 | gapdh | -0.061 |
| mrto4 | 0.112 | CR855257.1 | -0.060 |
| pfdn4 | 0.111 | slc25a4 | -0.059 |
| exosc3 | 0.111 | cavin4a | -0.059 |
| cct2 | 0.111 | sgcd | -0.059 |
| myog | 0.110 | tnni2a.4 | -0.059 |