methionine sulfoxide reductase B3
ZFIN 
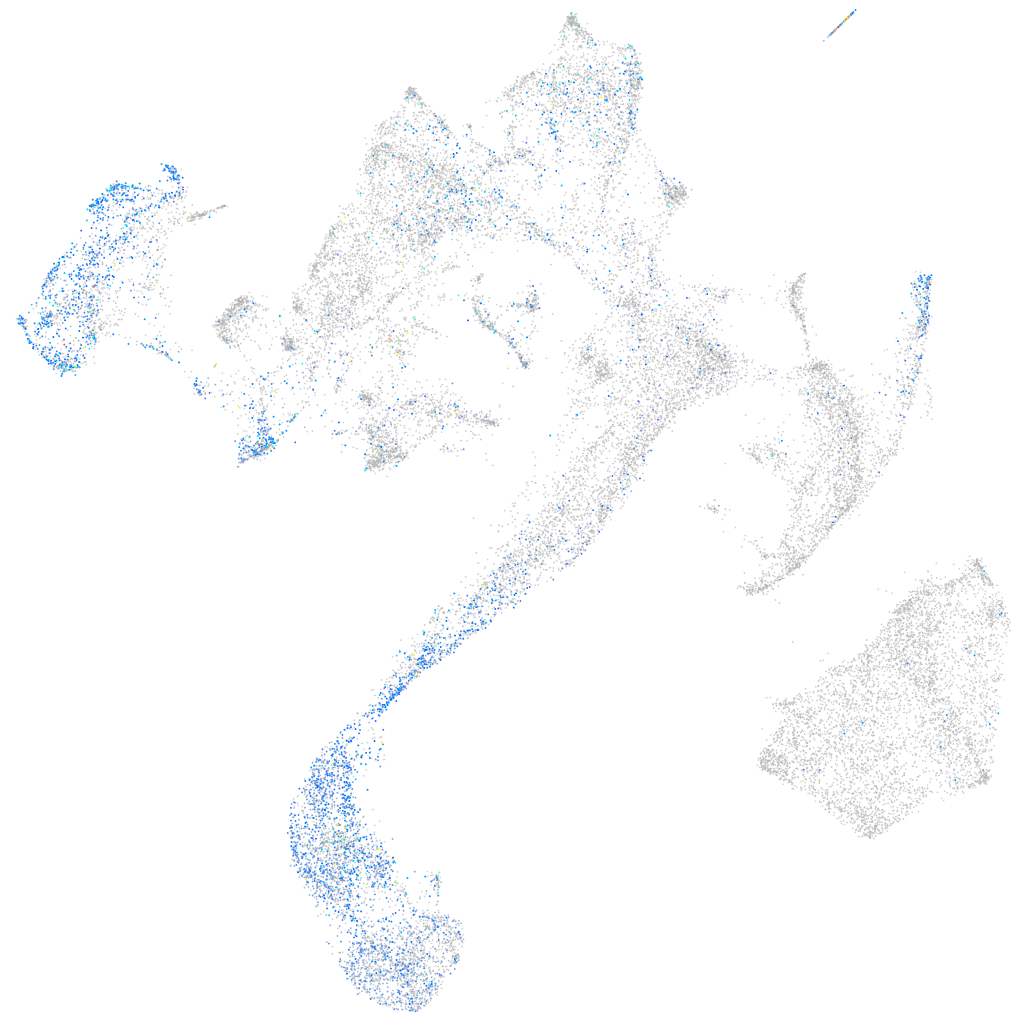
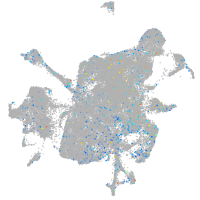
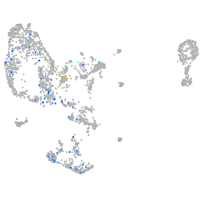
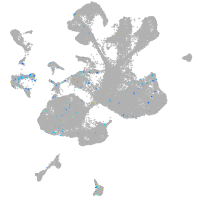
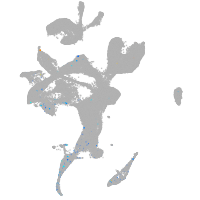
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| casq2 | 0.416 | hsp90ab1 | -0.361 |
| cavin4b | 0.400 | pabpc1a | -0.352 |
| neb | 0.397 | h3f3d | -0.339 |
| aldoab | 0.393 | hmgb2b | -0.332 |
| ldb3b | 0.392 | si:ch73-1a9.3 | -0.330 |
| CABZ01072309.1 | 0.391 | ran | -0.330 |
| atp2a1 | 0.391 | hnrnpaba | -0.328 |
| gapdh | 0.390 | khdrbs1a | -0.326 |
| ckma | 0.389 | h2afvb | -0.326 |
| srl | 0.389 | hnrnpabb | -0.324 |
| ak1 | 0.387 | cirbpb | -0.322 |
| ckmb | 0.387 | cirbpa | -0.319 |
| cav3 | 0.386 | setb | -0.307 |
| ldb3a | 0.384 | si:ch211-222l21.1 | -0.307 |
| myom1a | 0.383 | cbx3a | -0.306 |
| actn3b | 0.382 | hmgb2a | -0.306 |
| eno3 | 0.381 | ptmab | -0.305 |
| si:ch211-266g18.10 | 0.381 | hmga1a | -0.304 |
| actn3a | 0.380 | hnrnpa0b | -0.301 |
| pgam2 | 0.377 | si:ch73-281n10.2 | -0.301 |
| trdn | 0.376 | syncrip | -0.297 |
| tmem38a | 0.375 | hmgn2 | -0.293 |
| tnnc2 | 0.375 | snrpf | -0.291 |
| actc1b | 0.373 | hmgn7 | -0.290 |
| ttn.2 | 0.373 | npm1a | -0.286 |
| acta1b | 0.370 | anp32b | -0.284 |
| zgc:158296 | 0.369 | nop58 | -0.284 |
| ank1a | 0.369 | ncl | -0.282 |
| cavin4a | 0.368 | ubc | -0.280 |
| tmod4 | 0.367 | nop56 | -0.278 |
| tmem182a | 0.366 | snrpb | -0.278 |
| smyd1a | 0.366 | seta | -0.277 |
| desma | 0.365 | snrpd1 | -0.277 |
| rtn2a | 0.364 | tuba8l4 | -0.275 |
| tnnt3a | 0.363 | snu13b | -0.274 |