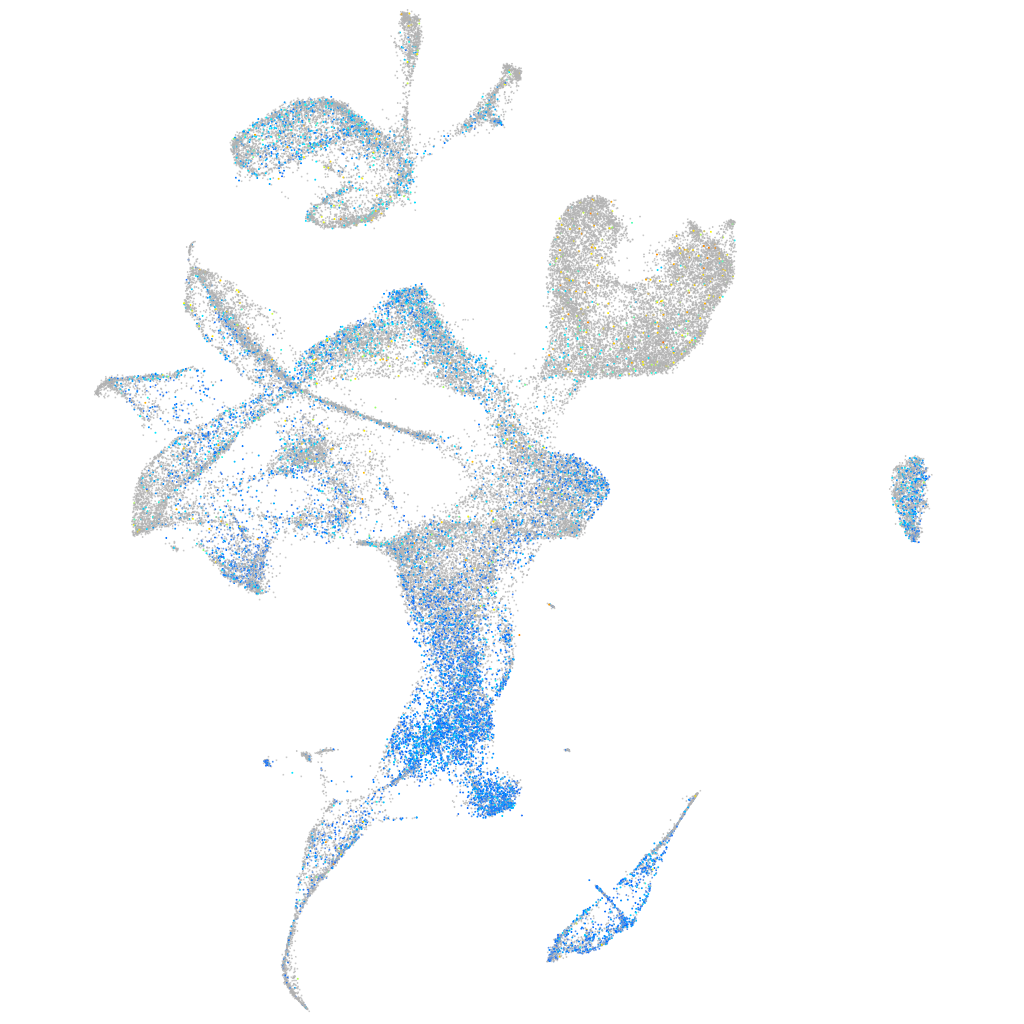
mitochondrial ribosomal protein L9
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| snu13b | 0.263 | ckbb | -0.148 |
| npm1a | 0.256 | CR383676.1 | -0.136 |
| nop58 | 0.248 | gpm6aa | -0.121 |
| nop10 | 0.248 | vsx1 | -0.119 |
| nhp2 | 0.239 | gpm6ab | -0.114 |
| dkc1 | 0.235 | ndrg4 | -0.114 |
| hspe1 | 0.234 | syt5b | -0.111 |
| c1qbp | 0.232 | calb2b | -0.110 |
| nop56 | 0.231 | gng13b | -0.109 |
| pa2g4a | 0.226 | ptmaa | -0.103 |
| rpl7l1 | 0.223 | snap25b | -0.103 |
| si:dkey-102m7.3 | 0.222 | gnb3a | -0.101 |
| fbl | 0.218 | mdkb | -0.101 |
| nifk | 0.218 | pvalb2 | -0.100 |
| tomm20a | 0.218 | epb41a | -0.099 |
| nop2 | 0.214 | pvalb1 | -0.099 |
| hspd1 | 0.213 | neurod4 | -0.098 |
| bysl | 0.211 | mpp6b | -0.097 |
| nip7 | 0.210 | ptmab | -0.097 |
| paics | 0.210 | nova2 | -0.097 |
| wdr43 | 0.210 | scrt2 | -0.096 |
| eif6 | 0.210 | samsn1a | -0.094 |
| mrto4 | 0.209 | btg1 | -0.093 |
| si:ch211-217k17.7 | 0.208 | lin7a | -0.093 |
| rcl1 | 0.207 | gnao1b | -0.093 |
| rsl1d1 | 0.206 | histh1l | -0.093 |
| ncl | 0.205 | rnasekb | -0.092 |
| rrp1 | 0.205 | isl1 | -0.092 |
| npm3 | 0.205 | pcp4l1 | -0.091 |
| bxdc2 | 0.201 | atp6v0cb | -0.090 |
| ssb | 0.201 | vamp1 | -0.090 |
| prps1a | 0.200 | nrn1lb | -0.090 |
| ppan | 0.198 | ccng2 | -0.087 |
| ebna1bp2 | 0.198 | rtn1a | -0.086 |
| mrpl12 | 0.196 | actc1b | -0.086 |