mitochondrial ribosomal protein L54
ZFIN 
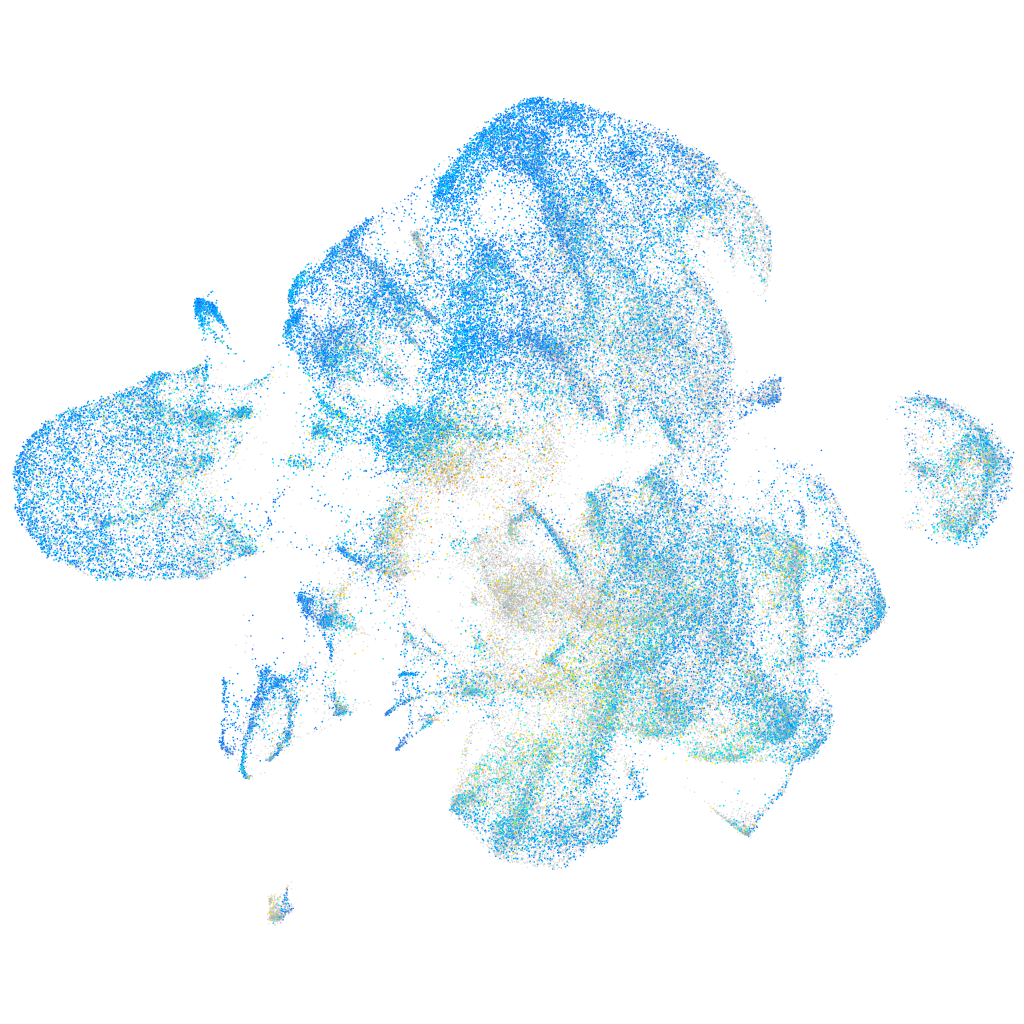
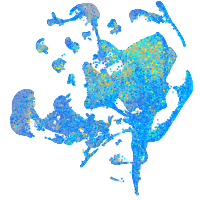
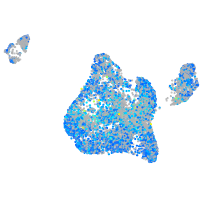
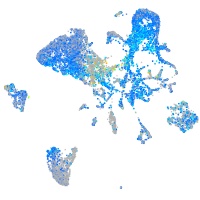
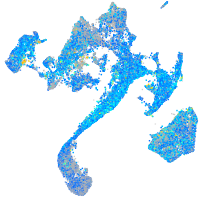
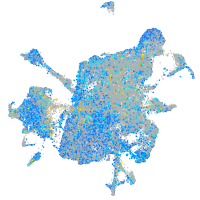
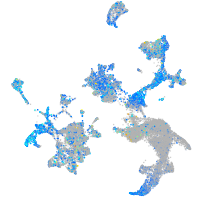
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| snu13b | 0.235 | CR383676.1 | -0.141 |
| npm1a | 0.231 | elavl3 | -0.132 |
| nhp2 | 0.218 | rtn1a | -0.126 |
| nop10 | 0.215 | gpm6aa | -0.125 |
| nop58 | 0.215 | COX3 | -0.120 |
| dkc1 | 0.204 | myt1b | -0.106 |
| cnbpa | 0.197 | nova2 | -0.102 |
| nop56 | 0.197 | mt-co2 | -0.100 |
| c1qbp | 0.196 | gpm6ab | -0.092 |
| rpl7l1 | 0.196 | stmn1b | -0.090 |
| nifk | 0.195 | ptmaa | -0.084 |
| hspe1 | 0.188 | myt1a | -0.084 |
| ranbp1 | 0.188 | actc1b | -0.081 |
| pa2g4a | 0.183 | atp6v0cb | -0.080 |
| nip7 | 0.183 | LOC100537384 | -0.078 |
| fbl | 0.183 | tp53inp1 | -0.077 |
| ncl | 0.183 | pvalb2 | -0.077 |
| si:ch211-217k17.7 | 0.183 | NC-002333.17 | -0.076 |
| cbx3a | 0.181 | dlb | -0.076 |
| paics | 0.179 | rnasekb | -0.076 |
| setb | 0.178 | tuba1c | -0.075 |
| nop2 | 0.177 | pvalb1 | -0.075 |
| rcl1 | 0.176 | tmsb | -0.073 |
| ptges3b | 0.176 | insm1a | -0.072 |
| bysl | 0.176 | tubb5 | -0.072 |
| mrto4 | 0.175 | aplp1 | -0.072 |
| si:dkey-102m7.3 | 0.175 | hbbe1.3 | -0.071 |
| wdr43 | 0.174 | LOC798783 | -0.071 |
| rsl1d1 | 0.174 | cspg5a | -0.071 |
| mak16 | 0.171 | mt-co1 | -0.071 |
| bxdc2 | 0.170 | nsg2 | -0.070 |
| etf1b | 0.170 | appa | -0.069 |
| snrpd1 | 0.170 | vamp2 | -0.069 |
| anp32b | 0.169 | serinc1 | -0.069 |
| selenoh | 0.168 | hbae3 | -0.069 |