myosin phosphatase Rho interacting protein
ZFIN 
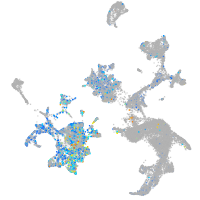
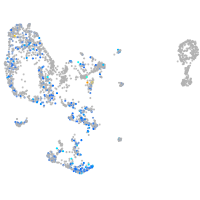
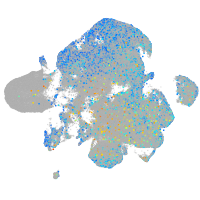
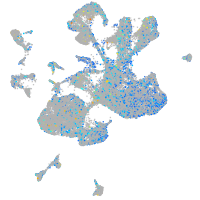
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tubb2b | 0.073 | fabp7a | -0.047 |
| hnrnpaba | 0.071 | atp1a1b | -0.044 |
| nucks1a | 0.070 | si:ch211-66e2.5 | -0.043 |
| ilf2 | 0.067 | ptn | -0.041 |
| smarca4a | 0.067 | dap1b | -0.040 |
| hspa8 | 0.066 | cox4i2 | -0.037 |
| khdrbs1a | 0.066 | glula | -0.036 |
| cirbpa | 0.066 | slc1a2b | -0.035 |
| syncrip | 0.065 | pvalb2 | -0.035 |
| hnrnpa0b | 0.064 | apoa2 | -0.034 |
| hnrnpabb | 0.064 | cebpd | -0.034 |
| seta | 0.063 | slc6a1b | -0.034 |
| psmd8 | 0.061 | pvalb1 | -0.034 |
| atrx | 0.060 | slc1a3b | -0.033 |
| sumo3a | 0.060 | ppap2d | -0.033 |
| hnrnpa0l | 0.060 | cx43 | -0.033 |
| rbm4.3 | 0.059 | gngt2b | -0.033 |
| eef2b | 0.059 | nog2 | -0.032 |
| hnrnpa1a | 0.059 | zgc:165461 | -0.031 |
| cbx5 | 0.059 | cd63 | -0.031 |
| fkbp1aa | 0.058 | hspb15 | -0.031 |
| lin28a | 0.057 | ca4a | -0.030 |
| idh2 | 0.056 | mfge8a | -0.030 |
| ptmab | 0.056 | mdkb | -0.030 |
| si:ch211-51e12.7 | 0.056 | qki2 | -0.030 |
| marcksb | 0.056 | sept8b | -0.029 |
| hnrnpa0a | 0.056 | gpr37l1b | -0.029 |
| tubb5 | 0.056 | slc4a4a | -0.029 |
| tcp1 | 0.055 | aqp1a.1 | -0.029 |
| psmb7 | 0.055 | hbbe1.1 | -0.029 |
| si:ch211-288g17.3 | 0.054 | slc3a2a | -0.029 |
| smarce1 | 0.054 | tmem176 | -0.029 |
| coro1cb | 0.054 | ptgdsb.2 | -0.028 |
| eif3ea | 0.054 | apoa1b | -0.028 |
| hnrnpub | 0.053 | CU467822.1 | -0.028 |