"Moloney leukemia virus 10b, tandem duplicate 1"
ZFIN 
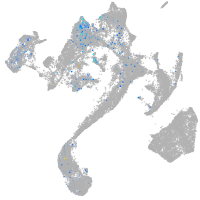
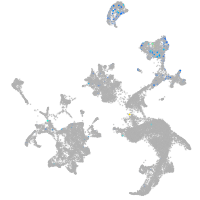
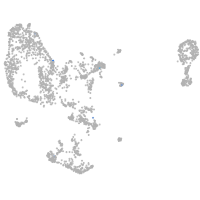
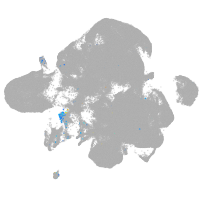
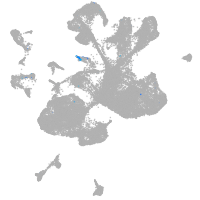
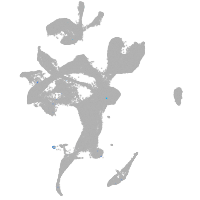
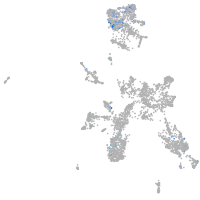
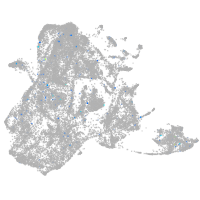
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX294160.1 | 0.841 | marcksl1b | -0.262 |
| lactbl1b | 0.810 | hnrnpaba | -0.255 |
| si:ch211-214j8.12 | 0.756 | si:ch73-281n10.2 | -0.253 |
| slc4a11 | 0.756 | ncl | -0.244 |
| pip5k1ba | 0.716 | sfpq | -0.239 |
| satb1a | 0.681 | anp32e | -0.238 |
| LOC103909808 | 0.650 | hmgb2a | -0.234 |
| triob | 0.620 | fbl | -0.232 |
| ela2 | 0.615 | NC-002333.4 | -0.232 |
| hoxc11a | 0.615 | nucks1a | -0.225 |
| LOC100536045 | 0.615 | serbp1b | -0.216 |
| si:dkey-220k22.1 | 0.615 | gpkow | -0.215 |
| socs1b | 0.615 | cdk11b | -0.214 |
| camk2n2 | 0.608 | srrt | -0.213 |
| AL928866.1 | 0.608 | h1m | -0.212 |
| CABZ01117752.1 | 0.607 | stm | -0.209 |
| lamb4 | 0.604 | ddit4 | -0.204 |
| LOC101886933 | 0.604 | hmga1a | -0.202 |
| si:zfos-364h11.2 | 0.604 | acin1b | -0.198 |
| BX664618.1 | 0.603 | cxcr4b | -0.198 |
| anxa1c | 0.594 | tbx16 | -0.197 |
| ank3a | 0.593 | snrpa | -0.197 |
| dgat1b | 0.590 | u2surp | -0.196 |
| AL929217.1 | 0.581 | akap17a | -0.196 |
| edaradd | 0.576 | pttg1 | -0.194 |
| limch1b | 0.575 | akap12b | -0.194 |
| ano9b | 0.566 | hmgb1a | -0.193 |
| trim35-3 | 0.566 | knop1 | -0.190 |
| slc18b1 | 0.565 | nono | -0.189 |
| fam149a | 0.552 | tpx2 | -0.187 |
| pik3c2g | 0.548 | ca15b | -0.185 |
| ubtd1a | 0.548 | rbm4.3 | -0.182 |
| LOC108181606 | 0.546 | ctsla | -0.181 |
| itfg1 | 0.545 | dnajc1 | -0.181 |
| ampd2b | 0.534 | hnrnpa1b | -0.179 |