matrix metallopeptidase 14a (membrane-inserted)
ZFIN 
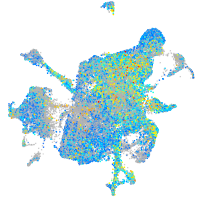
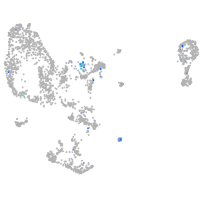
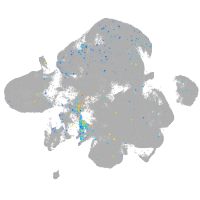
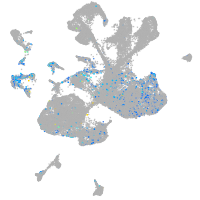
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc22a2 | 0.788 | npm1a | -0.366 |
| cables2b | 0.594 | nop58 | -0.321 |
| si:dkeyp-67a8.4 | 0.594 | hdgfl2 | -0.314 |
| bahcc1b | 0.581 | fbl | -0.295 |
| hic1 | 0.580 | u2surp | -0.288 |
| atg2b | 0.579 | hnrnpl2 | -0.287 |
| hoxa3a | 0.559 | cd2bp2 | -0.285 |
| herc3 | 0.546 | dkc1 | -0.281 |
| CABZ01068362.1 | 0.541 | epcam | -0.270 |
| CU457778.3 | 0.541 | zgc:56676 | -0.257 |
| en2a | 0.541 | ptmab | -0.256 |
| evplb | 0.541 | knop1 | -0.255 |
| grpr | 0.541 | brd7 | -0.245 |
| lgi2a | 0.541 | marcksb | -0.244 |
| lrrc7 | 0.541 | cdk11b | -0.243 |
| ppm1lb | 0.541 | nob1 | -0.238 |
| hoxb5b | 0.538 | rps6 | -0.235 |
| nadkb | 0.530 | metap2b | -0.232 |
| rgs12a | 0.526 | zgc:173742 | -0.232 |
| CR751227.2 | 0.522 | cwf19l2 | -0.230 |
| ext2 | 0.514 | prkrip1 | -0.229 |
| scarb2c | 0.512 | fam50a | -0.223 |
| gcga | 0.510 | cactin | -0.220 |
| mnd1 | 0.507 | rps18 | -0.220 |
| hoxd4a | 0.502 | ccnb1 | -0.212 |
| ehbp1l1a | 0.500 | si:ch211-161h7.4 | -0.212 |
| slc35c1 | 0.498 | apoc1 | -0.210 |
| hoxb2a | 0.496 | kpna2 | -0.209 |
| hoxb3a | 0.494 | nop56 | -0.208 |
| lman2la | 0.491 | dnttip2 | -0.207 |
| XLOC-031060 | 0.486 | pcna | -0.206 |
| CR382326.1 | 0.486 | top1l | -0.205 |
| hoxd3a | 0.481 | ebna1bp2 | -0.203 |
| zgc:174863 | 0.479 | dmap1 | -0.200 |
| dock7 | 0.474 | mphosph10 | -0.199 |