MLLT11 transcription factor 7 cofactor
ZFIN 
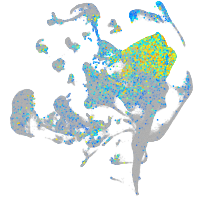
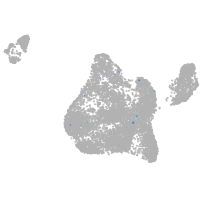
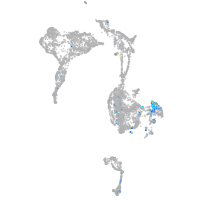
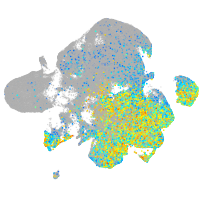
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tmsb | 0.338 | hdlbpa | -0.118 |
| scg3 | 0.337 | gapdh | -0.113 |
| neurod1 | 0.328 | aldh6a1 | -0.108 |
| gnao1a | 0.314 | gstr | -0.107 |
| vamp2 | 0.310 | ahcy | -0.104 |
| pcsk1nl | 0.307 | nupr1b | -0.103 |
| mir7a-1 | 0.302 | rpl4 | -0.101 |
| id4 | 0.301 | fbp1b | -0.099 |
| syt1a | 0.298 | cx32.3 | -0.097 |
| dpysl2b | 0.296 | gamt | -0.096 |
| atp6v0cb | 0.292 | aldh7a1 | -0.094 |
| rnasekb | 0.290 | aldob | -0.093 |
| gap43 | 0.286 | gstt1a | -0.091 |
| insm1b | 0.284 | glud1b | -0.090 |
| stxbp1a | 0.283 | slc16a1b | -0.089 |
| pax4 | 0.279 | adka | -0.088 |
| gdi1 | 0.275 | gatm | -0.088 |
| tuba1c | 0.275 | rps18 | -0.087 |
| snap25a | 0.273 | pnp4b | -0.087 |
| cnrip1a | 0.271 | eef1da | -0.086 |
| tspan7b | 0.269 | g6pca.2 | -0.086 |
| vat1 | 0.267 | ugt1a7 | -0.086 |
| stmn1b | 0.267 | mgst1.2 | -0.086 |
| egr4 | 0.266 | cyp4v8 | -0.085 |
| pax6b | 0.266 | agxta | -0.084 |
| si:ch211-255i3.4 | 0.265 | cpn1 | -0.084 |
| sncb | 0.263 | scp2a | -0.084 |
| gpc1a | 0.262 | shmt1 | -0.083 |
| zgc:65894 | 0.259 | aqp12 | -0.083 |
| gng3 | 0.256 | dhrs9 | -0.083 |
| fev | 0.254 | nipsnap3a | -0.082 |
| c2cd4a | 0.250 | apoa4b.1 | -0.082 |
| insm1a | 0.249 | sult2st2 | -0.081 |
| zgc:101731 | 0.248 | mat1a | -0.081 |
| rasd4 | 0.247 | haao | -0.081 |