"microtubule associated monooxygenase, calponin and LIM domain containing 3b"
ZFIN 
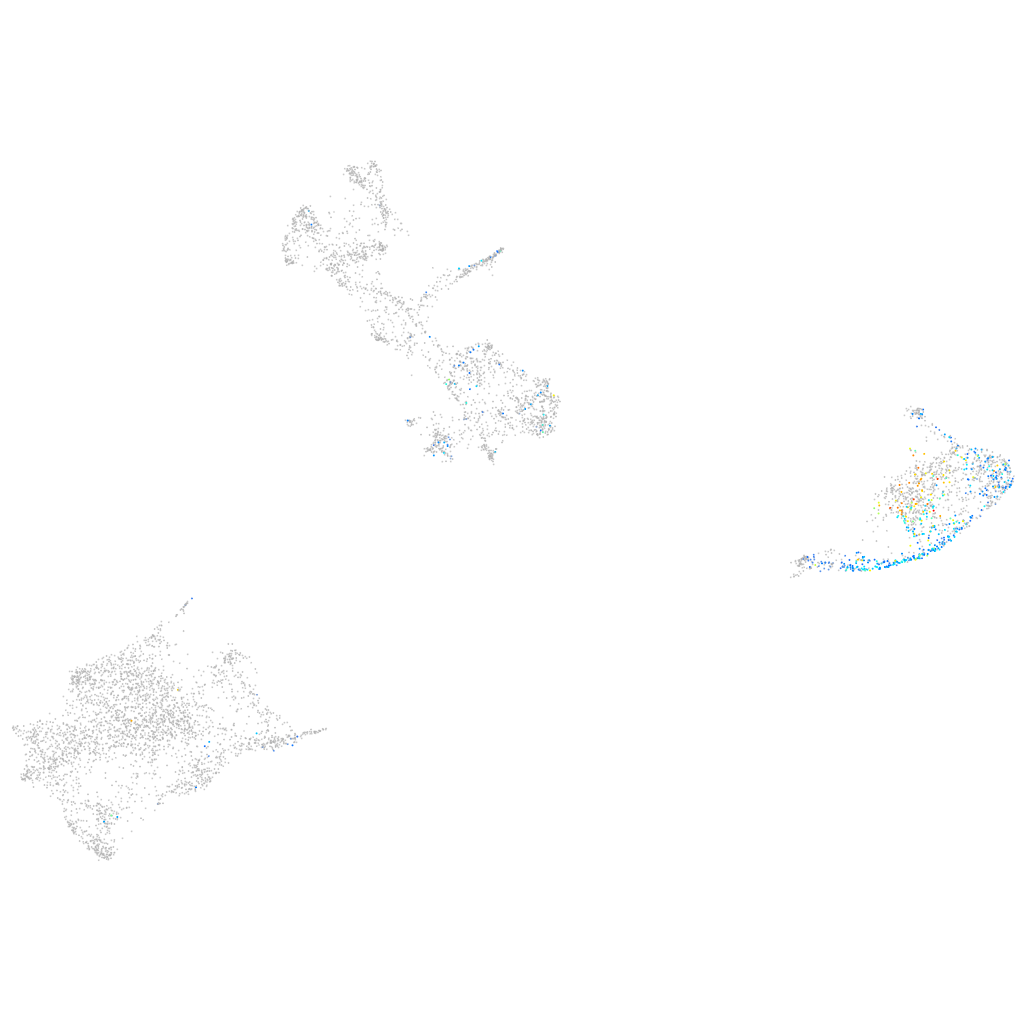
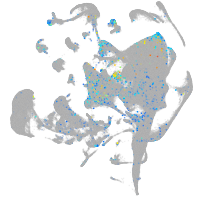
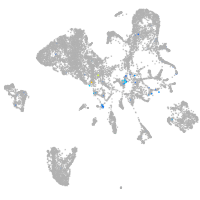
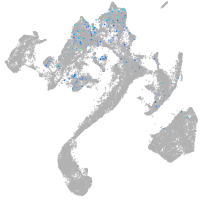
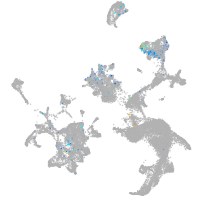
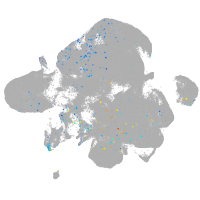
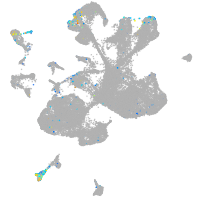
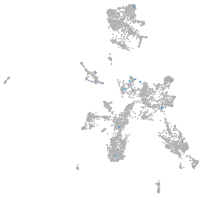
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dct | 0.415 | CABZ01021592.1 | -0.228 |
| zgc:91968 | 0.409 | paics | -0.221 |
| tyrp1b | 0.400 | uraha | -0.195 |
| si:ch73-389b16.1 | 0.398 | si:dkey-251i10.2 | -0.190 |
| pmela | 0.385 | tmem130 | -0.174 |
| tyrp1a | 0.381 | mdh1aa | -0.174 |
| slc24a5 | 0.364 | slc2a15a | -0.166 |
| tyr | 0.349 | phyhd1 | -0.165 |
| oca2 | 0.341 | aox5 | -0.148 |
| slc45a2 | 0.332 | glulb | -0.148 |
| gstt1a | 0.307 | atic | -0.147 |
| mtbl | 0.302 | si:ch211-251b21.1 | -0.147 |
| tmem243b | 0.296 | impdh1b | -0.144 |
| prkar1b | 0.294 | bscl2l | -0.131 |
| slc37a2 | 0.292 | aqp3a | -0.130 |
| kita | 0.286 | slc22a7a | -0.128 |
| rabl6b | 0.283 | gmps | -0.128 |
| tspan10 | 0.281 | prps1a | -0.127 |
| aadac | 0.280 | cyb5a | -0.126 |
| lamp1a | 0.277 | pax7b | -0.125 |
| anxa1a | 0.270 | pax7a | -0.120 |
| mitfa | 0.265 | bco1 | -0.117 |
| slc3a2a | 0.265 | prdx1 | -0.117 |
| tspan36 | 0.264 | sult1st1 | -0.116 |
| atp6v0ca | 0.261 | sprb | -0.115 |
| qdpra | 0.256 | cx30.3 | -0.112 |
| rab38 | 0.247 | TMEM19 | -0.112 |
| tfap2e | 0.245 | krt18b | -0.112 |
| mlpha | 0.245 | aldob | -0.107 |
| atp6v0a2b | 0.245 | prdx5 | -0.106 |
| pknox1.2 | 0.245 | pmp22b | -0.105 |
| tmem110l | 0.242 | ppat | -0.105 |
| si:dkeyp-69c1.9 | 0.241 | defbl1 | -0.104 |
| msx1b | 0.241 | oacyl | -0.103 |
| dync1li2 | 0.241 | rbp4l | -0.102 |