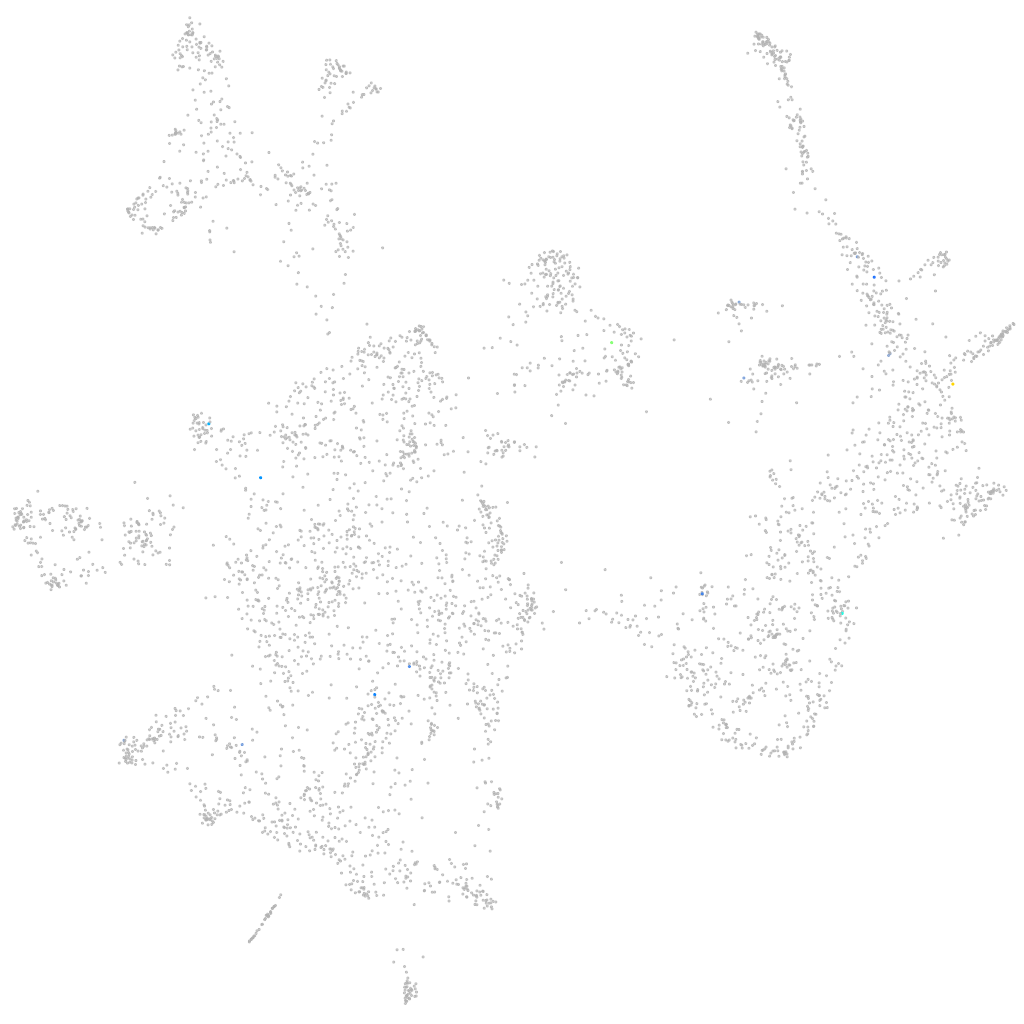
major histocompatibility complex class I UKA
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110439443 | 0.459 | rpl4 | -0.040 |
| diras1b | 0.421 | mt-cyb | -0.037 |
| neto1 | 0.389 | mt-nd2 | -0.036 |
| LOC110439775 | 0.349 | atp5mc1 | -0.033 |
| si:dkey-256e7.8 | 0.308 | hmgb1a | -0.033 |
| adgrd1 | 0.305 | mt-nd1 | -0.033 |
| chad | 0.304 | rab2a | -0.031 |
| gucy2f | 0.297 | mt-atp6 | -0.031 |
| zdhhc21.1 | 0.292 | csnk2b | -0.030 |
| gata5 | 0.286 | ndufa4l | -0.030 |
| trim35-12 | 0.278 | supt4h1 | -0.028 |
| si:dkey-57k2.6 | 0.267 | calm2b | -0.028 |
| arhgap20b | 0.261 | ndufb7 | -0.028 |
| tcf21 | 0.253 | NDUFB1 | -0.028 |
| mir725 | 0.249 | mt-co2 | -0.027 |
| wu:fc23c09 | 0.247 | nkap | -0.027 |
| c1ql3b | 0.243 | ndufa12 | -0.026 |
| CR769772.3 | 0.243 | ptges3a | -0.026 |
| samd7 | 0.241 | LOC110439372 | -0.026 |
| fam113 | 0.226 | ssr2 | -0.026 |
| pbx3a | 0.221 | ilf3b | -0.026 |
| BX323812.2 | 0.216 | dynll1 | -0.026 |
| BX294383.2 | 0.208 | hspe1 | -0.025 |
| BX321875.3 | 0.208 | cap1 | -0.025 |
| si:dkey-265e7.1 | 0.206 | emc10 | -0.025 |
| si:ch211-217k17.10 | 0.200 | zmat2 | -0.025 |
| trim24 | 0.199 | polr2f | -0.025 |
| CR626944.1 | 0.197 | eif2s1b | -0.025 |
| adra2a | 0.194 | pdia6 | -0.025 |
| CR749162.1 | 0.192 | rab3ip | -0.025 |
| kcng2 | 0.191 | cdk2ap2 | -0.024 |
| CR848705.2 | 0.185 | ip6k2a | -0.024 |
| si:dkey-23k10.3 | 0.184 | ndufs5 | -0.024 |
| ca14 | 0.177 | zgc:92313 | -0.024 |
| zgc:64201 | 0.176 | STMP1 | -0.024 |