"alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase B"
ZFIN 
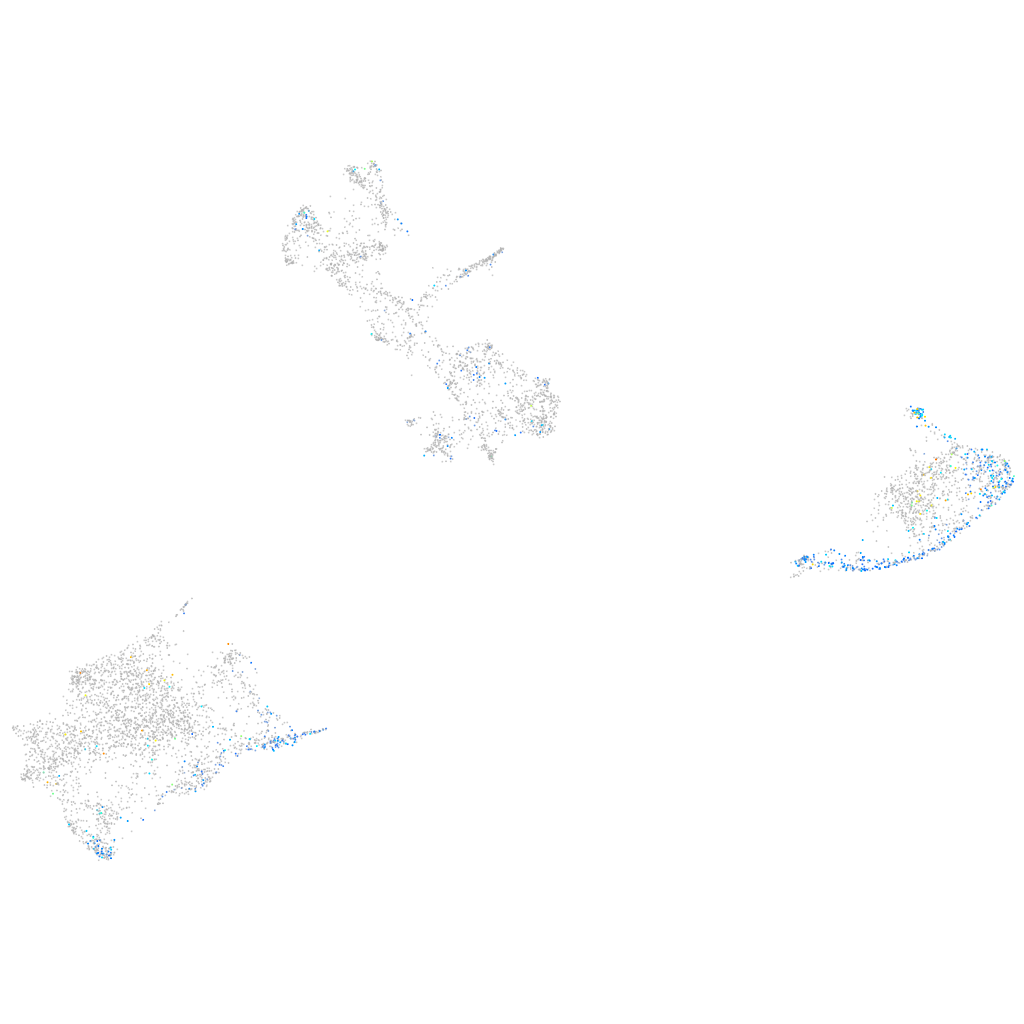
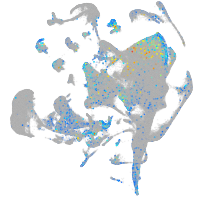
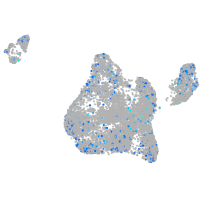
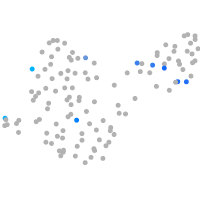
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kita | 0.231 | uraha | -0.141 |
| si:ch73-389b16.1 | 0.225 | CABZ01021592.1 | -0.114 |
| tfap2e | 0.224 | paics | -0.114 |
| tmem243b | 0.221 | si:dkey-251i10.2 | -0.108 |
| slc45a2 | 0.206 | mdh1aa | -0.106 |
| phlda1 | 0.205 | defbl1 | -0.094 |
| SPAG9 | 0.199 | apoda.1 | -0.088 |
| mitfa | 0.193 | hbae3 | -0.086 |
| lamp1a | 0.192 | hbbe1.3 | -0.085 |
| slc3a2a | 0.191 | mdkb | -0.083 |
| tspan36 | 0.191 | phyhd1 | -0.082 |
| pmela | 0.189 | slc2a15a | -0.081 |
| tyr | 0.189 | actc1b | -0.080 |
| slc24a5 | 0.186 | si:ch211-256m1.8 | -0.078 |
| rabl6b | 0.185 | gpnmb | -0.078 |
| tfap2a | 0.181 | krt4 | -0.077 |
| zgc:91968 | 0.178 | unm-sa821 | -0.076 |
| sypl2b | 0.177 | prdx1 | -0.076 |
| tyrp1b | 0.175 | lypc | -0.076 |
| oca2 | 0.175 | rbp4l | -0.075 |
| pah | 0.174 | pvalb2 | -0.075 |
| tyrp1a | 0.172 | pvalb1 | -0.074 |
| msx1b | 0.171 | si:dkey-197i20.6 | -0.072 |
| cdh1 | 0.171 | hmgn2 | -0.070 |
| slc7a5 | 0.170 | si:ch211-222l21.1 | -0.070 |
| mchr2 | 0.169 | tmem130 | -0.070 |
| si:zfos-943e10.1 | 0.168 | slc25a36a | -0.069 |
| dct | 0.167 | s100v2 | -0.068 |
| aadac | 0.163 | sult1st1 | -0.067 |
| pcdh10a | 0.161 | hbae1.1 | -0.067 |
| si:ch211-39i22.1 | 0.161 | impdh1b | -0.066 |
| tspan10 | 0.160 | si:ch211-251b21.1 | -0.065 |
| pcdh9 | 0.160 | atic | -0.065 |
| slc22a2 | 0.160 | ponzr1 | -0.065 |
| nr2f5 | 0.159 | tuba1c | -0.063 |