mex-3 RNA binding family member B
ZFIN 
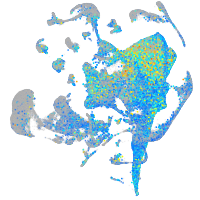
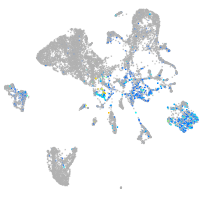
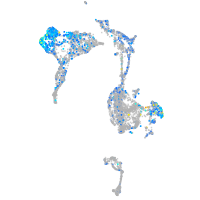
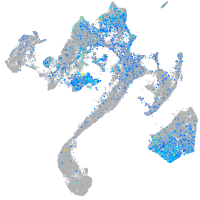
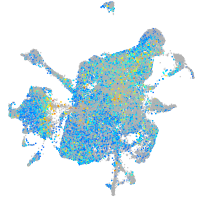
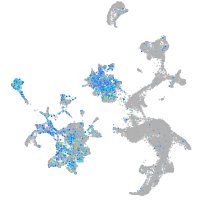
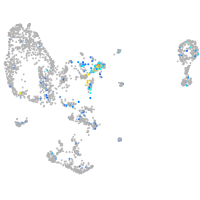
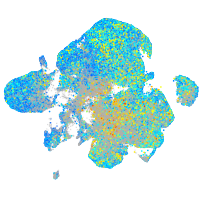
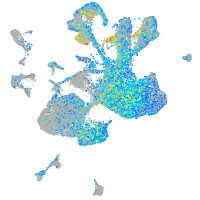
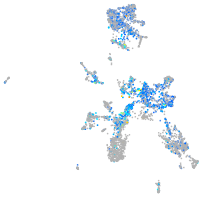
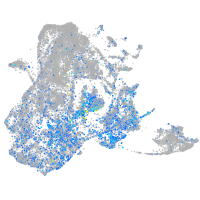
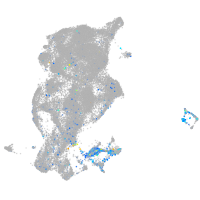
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.440 | cox7c | -0.170 |
| nova2 | 0.396 | atp5l | -0.167 |
| rtn1a | 0.382 | cdh17 | -0.158 |
| dpysl2b | 0.375 | rbm47 | -0.158 |
| gpm6aa | 0.360 | cox7b | -0.153 |
| FO082781.1 | 0.356 | cox6c | -0.152 |
| apc2 | 0.346 | atp5md | -0.151 |
| tmsb | 0.340 | COX5B | -0.148 |
| rbfox1 | 0.333 | elovl1b | -0.147 |
| tuba1c | 0.331 | atp1a1a.1 | -0.146 |
| celf2 | 0.330 | sgk1 | -0.145 |
| tubb5 | 0.329 | sypl2a | -0.143 |
| pbx3b | 0.320 | hnf1ba | -0.143 |
| onecut1 | 0.320 | suclg1 | -0.143 |
| si:dkey-276j7.1 | 0.308 | atp5mf | -0.142 |
| stmn1b | 0.306 | ezra | -0.142 |
| marcksl1a | 0.301 | cox7a2a | -0.141 |
| myt1a | 0.290 | ndrg1a | -0.141 |
| nsg2 | 0.289 | cdaa | -0.141 |
| myt1b | 0.287 | cox6b2 | -0.139 |
| nkain1 | 0.287 | uqcrfs1 | -0.138 |
| si:ch73-386h18.1 | 0.286 | ndufa4l | -0.137 |
| insm1a | 0.285 | slc43a2b | -0.136 |
| ckbb | 0.284 | atp5meb | -0.136 |
| chn1 | 0.279 | degs1 | -0.136 |
| scrt2 | 0.278 | atp5pf | -0.135 |
| tmeff1b | 0.277 | cox6a2 | -0.135 |
| scrt1a | 0.277 | cd164 | -0.134 |
| tuba1a | 0.275 | atp5if1a | -0.134 |
| csdc2a | 0.275 | slc2a12 | -0.134 |
| mab21l2 | 0.271 | gpx1a | -0.133 |
| elavl4 | 0.270 | cox7a1 | -0.133 |
| id4 | 0.269 | sgk2a | -0.132 |
| marcksl1b | 0.268 | cox4i1 | -0.132 |
| pbx1a | 0.264 | uqcrh | -0.131 |