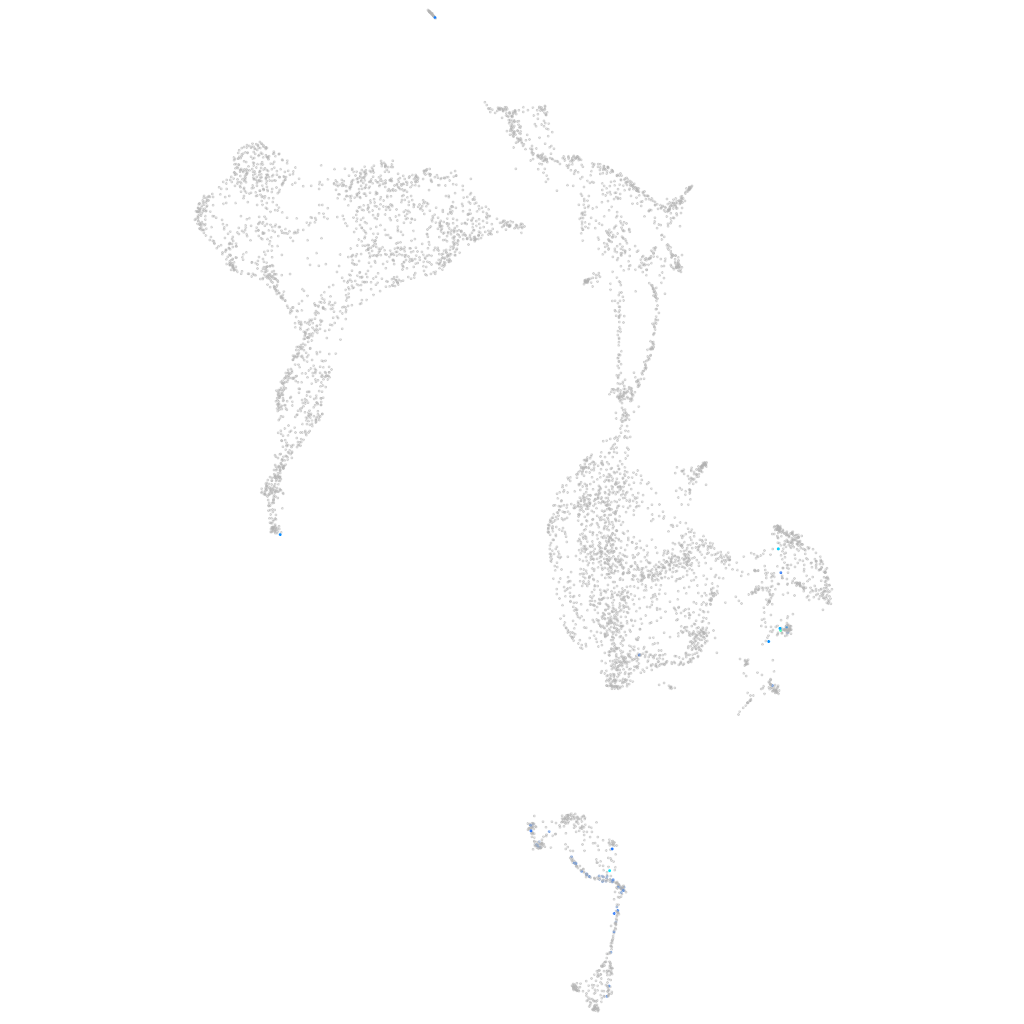
mesenchyme homeobox 2a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 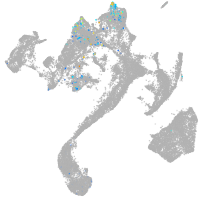 |
mural cells / non-skeletal muscle 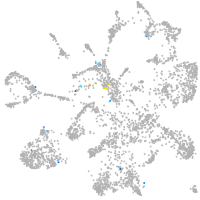 |
mesenchyme 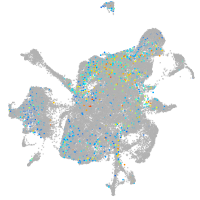 |
hematopoietic / vasculature 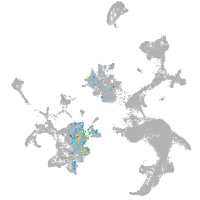 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis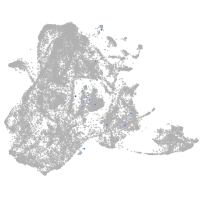 |
periderm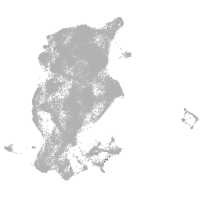 |
fin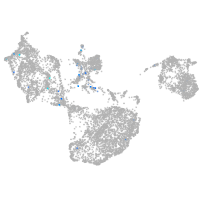 |
ionocytes / mucous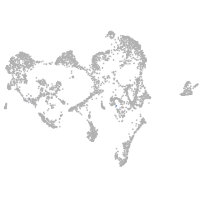 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkeyp-106c3.1 | 0.365 | ptmab | -0.087 |
| cyp1c2 | 0.355 | qkia | -0.084 |
| kcng1 | 0.353 | tbxta | -0.082 |
| gpr22b | 0.332 | h2afvb | -0.071 |
| or111-6 | 0.325 | cirbpa | -0.070 |
| FO704741.1 | 0.308 | anp32a | -0.069 |
| RF00263 | 0.269 | serpinh1b | -0.069 |
| apof | 0.263 | hmga1a | -0.068 |
| hmha1a | 0.243 | ctnnb1 | -0.067 |
| asip1 | 0.235 | dynll1 | -0.066 |
| nhsl1a | 0.232 | sept15 | -0.066 |
| rtn4rl2b | 0.228 | top1l | -0.065 |
| twist1b | 0.228 | p4ha2 | -0.064 |
| epob | 0.220 | hnrnpaba | -0.063 |
| si:ch211-202m22.1 | 0.219 | acin1a | -0.063 |
| si:dkeyp-33c10.7 | 0.219 | hnrnpabb | -0.063 |
| CU463231.2 | 0.219 | si:ch211-288g17.3 | -0.063 |
| lyve1a | 0.219 | hnrnpa0b | -0.062 |
| ftr80 | 0.218 | marcksl1b | -0.061 |
| LOC103911838 | 0.216 | shha | -0.061 |
| vegfd | 0.210 | plod1a | -0.060 |
| carmil3 | 0.204 | hmgn7 | -0.060 |
| pdgfra | 0.201 | cbx3a | -0.060 |
| cldn11a | 0.198 | snrnp70 | -0.060 |
| CU927890.1 | 0.194 | serf2 | -0.060 |
| flt4 | 0.193 | hmgb2b | -0.059 |
| c2cd4a | 0.193 | seta | -0.059 |
| rd3 | 0.191 | marcksb | -0.058 |
| pimr188 | 0.190 | hspb1 | -0.058 |
| olfml2bb | 0.189 | taf15 | -0.058 |
| s1pr3a | 0.189 | nucks1a | -0.058 |
| fli1b | 0.187 | rap1aa | -0.058 |
| LOC108182616 | 0.187 | khdrbs1a | -0.058 |
| si:dkey-151m22.8 | 0.187 | sf3b5 | -0.057 |
| ACKR2 | 0.186 | snrpb | -0.057 |