mediator complex subunit 24
ZFIN 
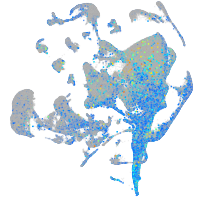
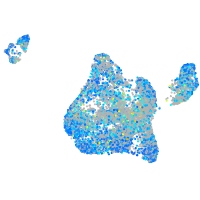
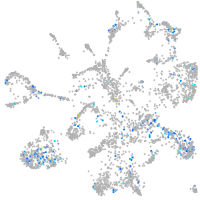
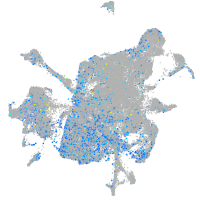
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgb2a | 0.155 | ckbb | -0.091 |
| hmgb2b | 0.155 | gapdhs | -0.090 |
| hmga1a | 0.150 | slc1a2b | -0.084 |
| h2afvb | 0.147 | acbd7 | -0.081 |
| hnrnpabb | 0.144 | gpm6ab | -0.080 |
| ran | 0.143 | CR383676.1 | -0.079 |
| snrpf | 0.142 | aldocb | -0.079 |
| snrpd1 | 0.141 | elavl4 | -0.079 |
| snrpb | 0.138 | glula | -0.077 |
| ddx39ab | 0.137 | cx43 | -0.075 |
| snrpe | 0.136 | calm1a | -0.074 |
| snrpg | 0.135 | snap25a | -0.073 |
| khdrbs1a | 0.134 | ppdpfb | -0.073 |
| hnrnpa0l | 0.128 | rnasekb | -0.072 |
| XLOC-003689 | 0.128 | pvalb2 | -0.071 |
| dut | 0.127 | pvalb1 | -0.070 |
| rps7 | 0.127 | ptn | -0.070 |
| zgc:56493 | 0.126 | gng3 | -0.070 |
| mki67 | 0.126 | ndrg3a | -0.069 |
| snrpd2 | 0.126 | fez1 | -0.066 |
| apex1 | 0.126 | slc4a4a | -0.066 |
| cbx3a | 0.125 | eno2 | -0.066 |
| seta | 0.125 | mt2 | -0.066 |
| eef1a1l1 | 0.123 | cplx2 | -0.066 |
| rplp0 | 0.123 | cadm4 | -0.066 |
| hnrnpaba | 0.122 | atp6v0cb | -0.065 |
| ccna2 | 0.122 | efhd1 | -0.065 |
| setb | 0.122 | sncgb | -0.065 |
| ddx39aa | 0.121 | sncb | -0.065 |
| ranbp1 | 0.121 | anxa13 | -0.065 |
| rps3a | 0.121 | vamp2 | -0.064 |
| h3f3a | 0.120 | si:ch211-66e2.5 | -0.064 |
| tuba8l4 | 0.120 | IGLON5 | -0.064 |
| si:ch211-222l21.1 | 0.120 | tpi1b | -0.064 |
| alyref | 0.120 | si:dkeyp-75h12.5 | -0.064 |