MAM domain containing glycosylphosphatidylinositol anchor 1
ZFIN 
Other cell groups


all cells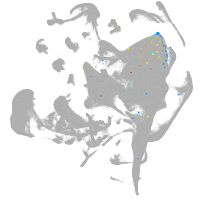 |
blastomeres |
PGCs |
endoderm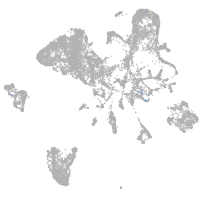 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural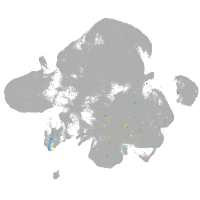 |
spinal cord / glia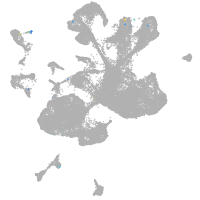 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| map1aa | 0.064 | aldob | -0.023 |
| ywhag2 | 0.064 | hspb1 | -0.020 |
| stmn2a | 0.063 | apoeb | -0.018 |
| zgc:65894 | 0.061 | epcam | -0.018 |
| slc17a6b | 0.059 | stm | -0.018 |
| elavl4 | 0.058 | apoc1 | -0.017 |
| snap25a | 0.058 | cx43.4 | -0.017 |
| tubb2 | 0.058 | eif4ebp3l | -0.017 |
| rtn1b | 0.057 | f11r.1 | -0.017 |
| nrn1a | 0.056 | mki67 | -0.017 |
| sncb | 0.056 | npm1a | -0.017 |
| sst6 | 0.056 | pou5f3 | -0.017 |
| tmsb2 | 0.056 | wu:fb18f06 | -0.017 |
| lgi2b | 0.055 | arf1 | -0.016 |
| si:dkey-43k4.5 | 0.055 | ccnd1 | -0.016 |
| stx1b | 0.055 | ccng1 | -0.016 |
| atp2b3b | 0.054 | cd9b | -0.016 |
| nrxn1a | 0.054 | cfl1l | -0.016 |
| zgc:153426 | 0.054 | dkc1 | -0.016 |
| gng3 | 0.053 | fbl | -0.016 |
| chga | 0.052 | nop58 | -0.016 |
| gap43 | 0.052 | polr3gla | -0.016 |
| nefma | 0.052 | s100a1 | -0.016 |
| tmem59l | 0.052 | spint2 | -0.016 |
| tuba2 | 0.052 | tpx2 | -0.016 |
| ntrk1 | 0.052 | ucp2 | -0.016 |
| gnao1a | 0.051 | zgc:56699 | -0.016 |
| mllt11 | 0.051 | asb11 | -0.015 |
| rab6bb | 0.051 | ccna2 | -0.015 |
| scg2b | 0.051 | ccnb1 | -0.015 |
| slc17a6a | 0.051 | cdh1 | -0.015 |
| ywhag1 | 0.051 | chaf1a | -0.015 |
| atp6v0cb | 0.050 | faub | -0.015 |
| map1ab | 0.050 | gapdh | -0.015 |
| map7d2b | 0.050 | gstp1 | -0.015 |




