MCF.2 cell line derived transforming sequence-like 2
ZFIN 
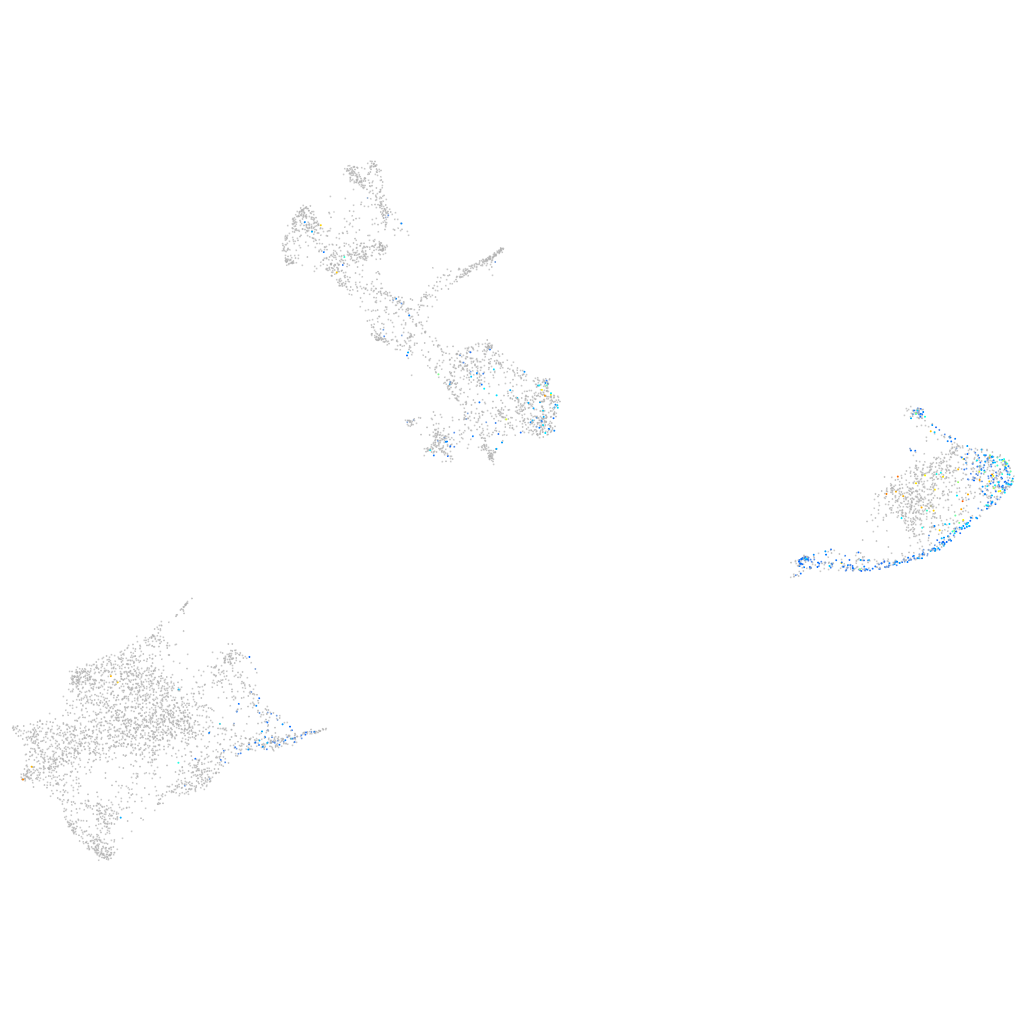
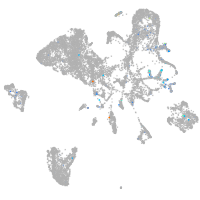
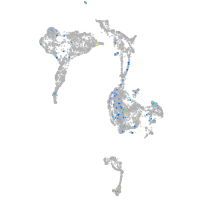
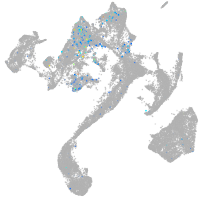
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.348 | uraha | -0.186 |
| kita | 0.334 | CABZ01021592.1 | -0.183 |
| slc24a5 | 0.293 | paics | -0.180 |
| pmela | 0.289 | mdh1aa | -0.163 |
| tyrp1b | 0.280 | si:dkey-251i10.2 | -0.159 |
| oca2 | 0.279 | tmem130 | -0.139 |
| slc7a5 | 0.275 | slc2a15a | -0.133 |
| tyrp1a | 0.272 | phyhd1 | -0.132 |
| dct | 0.271 | si:ch211-251b21.1 | -0.125 |
| slc39a10 | 0.265 | bscl2l | -0.113 |
| tyr | 0.264 | atic | -0.110 |
| aadac | 0.262 | impdh1b | -0.107 |
| lamp1a | 0.261 | cyb5a | -0.106 |
| si:ch73-389b16.1 | 0.254 | glulb | -0.105 |
| zgc:91968 | 0.251 | aox5 | -0.101 |
| tfap2e | 0.249 | krt18b | -0.099 |
| mchr2 | 0.247 | aldob | -0.099 |
| prkar1b | 0.246 | pmp22b | -0.097 |
| tfap2a | 0.246 | bco1 | -0.097 |
| slc22a2 | 0.244 | apoda.1 | -0.095 |
| gpr61 | 0.244 | defbl1 | -0.094 |
| rap1gap | 0.237 | prdx5 | -0.092 |
| kcnj13 | 0.237 | sult1st1 | -0.091 |
| megf10 | 0.234 | aqp3a | -0.090 |
| mitfa | 0.233 | TMEM19 | -0.090 |
| mtbl | 0.233 | oacyl | -0.090 |
| slc24a4a | 0.232 | pax7b | -0.089 |
| CDK18 | 0.229 | slc22a7a | -0.088 |
| slc30a1b | 0.229 | gpnmb | -0.087 |
| zdhhc2 | 0.227 | si:ch211-256m1.8 | -0.085 |
| slc30a8 | 0.225 | zgc:153031 | -0.084 |
| tmem243b | 0.222 | mthfd1b | -0.083 |
| slc45a2 | 0.221 | prps1a | -0.083 |
| slc6a15 | 0.220 | zgc:113337 | -0.083 |
| slc29a3 | 0.220 | lypc | -0.082 |