melanoma cell adhesion molecule b
ZFIN 
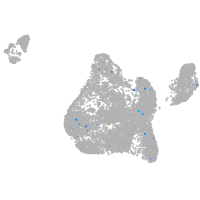
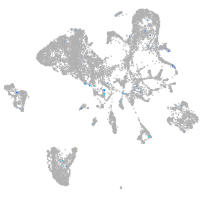
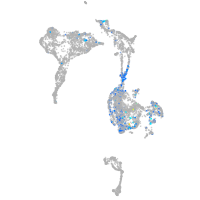
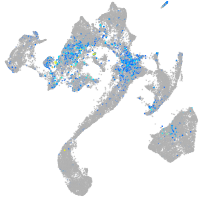
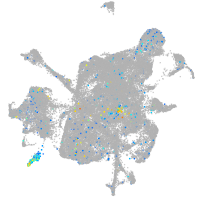
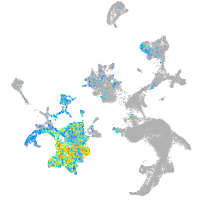
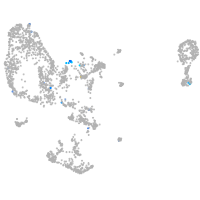
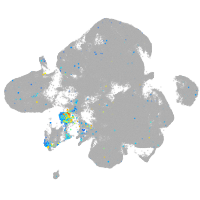
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scn4ab | 0.271 | gpm6aa | -0.028 |
| col4a2 | 0.215 | ptmab | -0.027 |
| col4a1 | 0.213 | cadm3 | -0.022 |
| pmp22a | 0.158 | si:ch73-1a9.3 | -0.021 |
| lamb1b | 0.153 | sinhcafl | -0.020 |
| tuba8l3 | 0.143 | neurod4 | -0.019 |
| phlda2 | 0.133 | hmgn6 | -0.019 |
| LOC108182616 | 0.132 | vsx1 | -0.018 |
| CT033796.1 | 0.131 | pax6a | -0.017 |
| BX088707.3 | 0.131 | rorb | -0.017 |
| lama4 | 0.131 | tmpob | -0.017 |
| itga1 | 0.129 | hnrnpaba | -0.017 |
| si:ch1073-291c23.2 | 0.129 | mpp6b | -0.017 |
| tmem176 | 0.128 | pax6b | -0.017 |
| col18a1b | 0.128 | calb2b | -0.016 |
| krt8 | 0.123 | nova2 | -0.016 |
| socs3a | 0.122 | ndrg4 | -0.016 |
| si:dkey-261h17.1 | 0.119 | sumo3a | -0.015 |
| ly6m5 | 0.116 | epb41a | -0.015 |
| krt18b | 0.116 | foxg1b | -0.015 |
| plvapb | 0.116 | smarce1 | -0.014 |
| etv2 | 0.115 | gng13b | -0.014 |
| foxd3 | 0.114 | scrt2 | -0.014 |
| agtr2 | 0.113 | pcbp3 | -0.014 |
| kdrl | 0.112 | marcksb | -0.014 |
| nkx2.3 | 0.111 | vsx2 | -0.013 |
| XLOC-043043 | 0.111 | cx43.4 | -0.013 |
| tgfbr2b | 0.110 | nfil3-2 | -0.013 |
| plvapa | 0.109 | h3f3d | -0.013 |
| marveld1 | 0.108 | hes6 | -0.013 |
| aoc2 | 0.107 | nrn1lb | -0.013 |
| BX322618.1 | 0.107 | yaf2 | -0.013 |
| si:ch211-171l17.4 | 0.107 | smarcd1 | -0.013 |
| erbb3b | 0.107 | ilf2 | -0.013 |
| inka1a | 0.107 | foxn4 | -0.013 |