mitogen-activated protein kinase 8 interacting protein 1a
ZFIN 
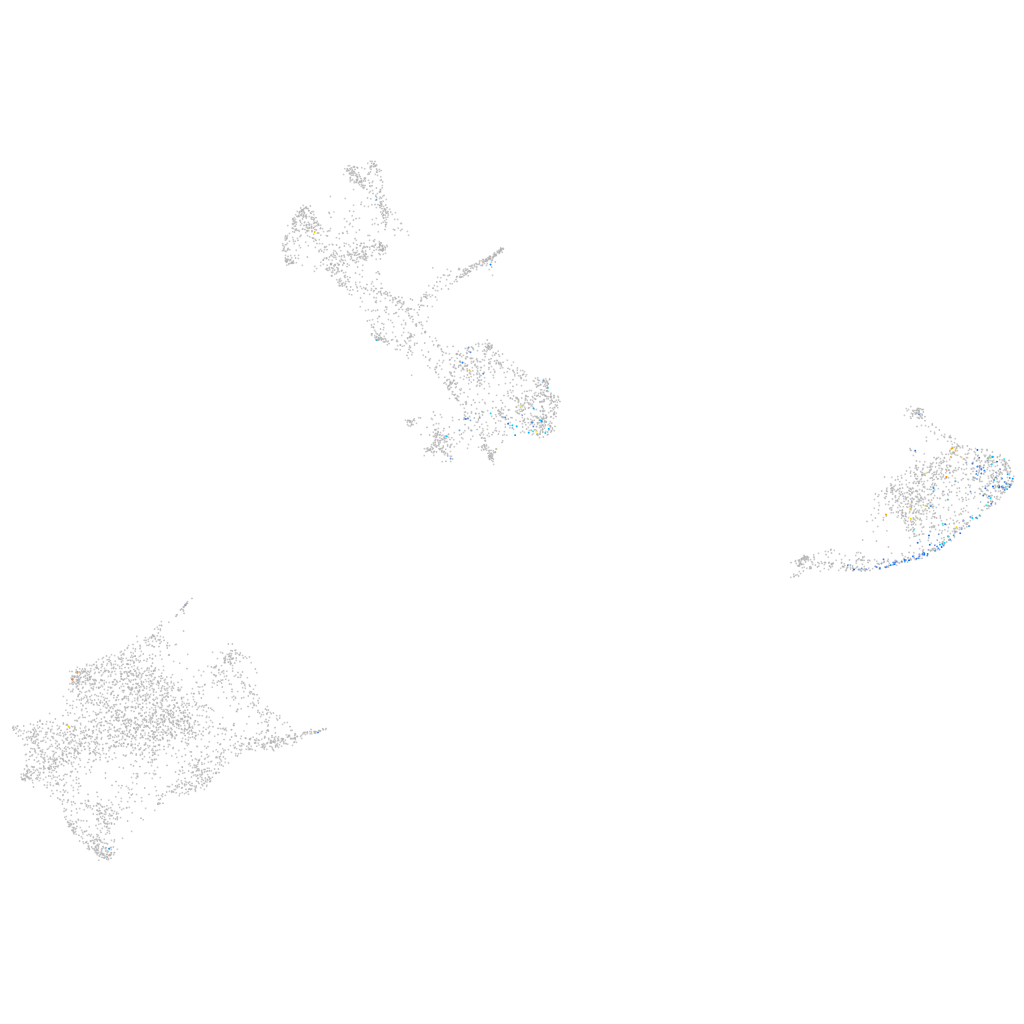
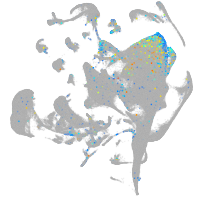
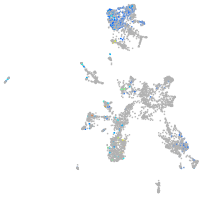
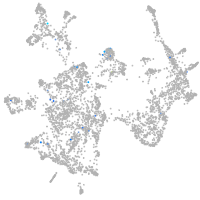
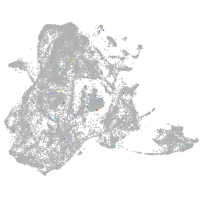
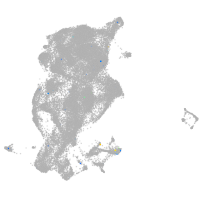
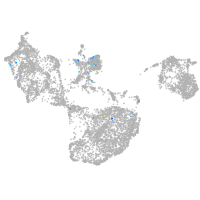
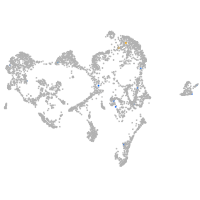
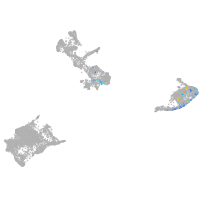
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| oca2 | 0.195 | paics | -0.118 |
| tyrp1b | 0.191 | CABZ01021592.1 | -0.104 |
| tyrp1a | 0.189 | mdh1aa | -0.100 |
| dct | 0.186 | si:dkey-251i10.2 | -0.094 |
| slc24a5 | 0.182 | atic | -0.092 |
| pmela | 0.182 | uraha | -0.092 |
| mtbl | 0.176 | glulb | -0.090 |
| prkar1b | 0.172 | impdh1b | -0.089 |
| zgc:91968 | 0.171 | tmem130 | -0.088 |
| kita | 0.170 | slc2a15a | -0.088 |
| si:ch73-389b16.1 | 0.168 | si:ch211-251b21.1 | -0.079 |
| slc39a10 | 0.168 | aox5 | -0.075 |
| CU466278.1 | 0.168 | bscl2l | -0.073 |
| slc22a2 | 0.164 | cyb5a | -0.073 |
| map7a | 0.163 | phyhd1 | -0.069 |
| tyr | 0.162 | bco1 | -0.068 |
| CDK18 | 0.156 | pax7a | -0.065 |
| aadac | 0.154 | ppat | -0.065 |
| slc30a1b | 0.154 | prdx1 | -0.065 |
| slc24a4a | 0.148 | gmps | -0.065 |
| SPAG9 | 0.148 | aldob | -0.065 |
| kcnj13 | 0.147 | pax7b | -0.065 |
| spra | 0.147 | sprb | -0.064 |
| CABZ01043506.1 | 0.145 | prps1a | -0.064 |
| XLOC-026713 | 0.143 | cax1 | -0.062 |
| rap1gap | 0.143 | si:ch211-194k22.8 | -0.062 |
| birc7 | 0.143 | sult1st1 | -0.059 |
| anxa1a | 0.142 | oacyl | -0.059 |
| slc7a5 | 0.142 | zgc:113337 | -0.059 |
| si:dkeyp-69c1.9 | 0.141 | shmt1 | -0.059 |
| pknox1.2 | 0.138 | krt18b | -0.058 |
| si:ch211-241j12.3 | 0.138 | rbp4l | -0.057 |
| slc29a3 | 0.138 | si:dkeyp-66d1.7 | -0.056 |
| LOC108180065 | 0.137 | mibp | -0.055 |
| gnao1a | 0.137 | cx30.3 | -0.054 |