mitogen-activated protein kinase 11
ZFIN 
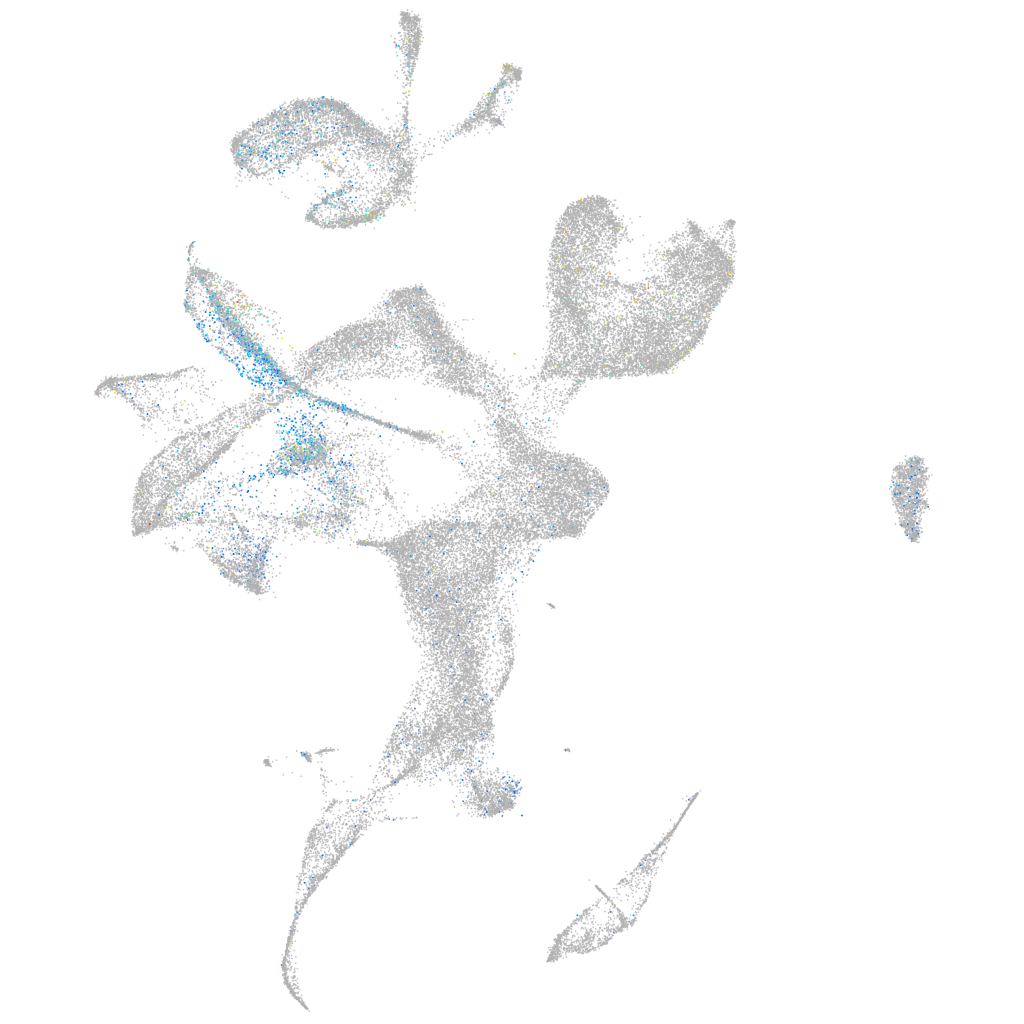
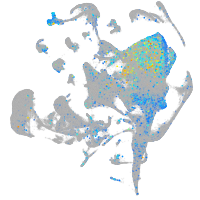
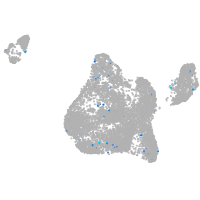
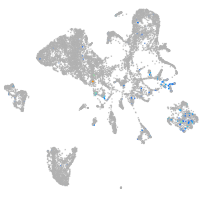
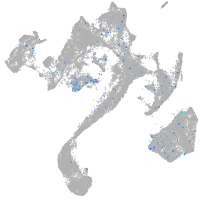
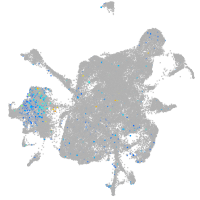
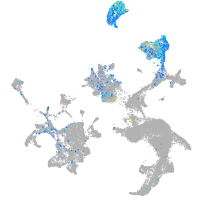
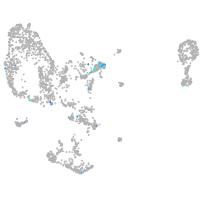
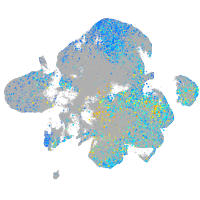
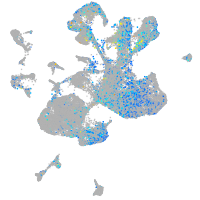
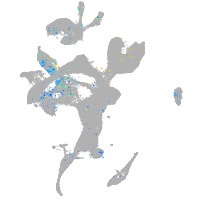
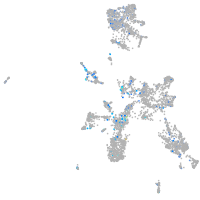
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gap43 | 0.229 | hmgb2a | -0.105 |
| tmsb | 0.212 | pcna | -0.061 |
| mllt11 | 0.207 | fabp7a | -0.061 |
| tubb5 | 0.201 | dut | -0.060 |
| stmn2a | 0.199 | msi1 | -0.060 |
| islr2 | 0.197 | stmn1a | -0.059 |
| rtn1b | 0.195 | rrm1 | -0.059 |
| elavl3 | 0.188 | lbr | -0.056 |
| stmn2b | 0.187 | mdka | -0.055 |
| dpysl3 | 0.179 | ccnd1 | -0.055 |
| ebf3a | 0.178 | mcm7 | -0.054 |
| rbpms2b | 0.177 | nutf2l | -0.053 |
| inab | 0.172 | selenoh | -0.053 |
| alcama | 0.172 | chaf1a | -0.053 |
| gng3 | 0.171 | ccna2 | -0.052 |
| nrn1a | 0.170 | anp32e | -0.052 |
| adcyap1b | 0.170 | dek | -0.052 |
| si:dkey-280e21.3 | 0.165 | mki67 | -0.051 |
| stmn1b | 0.161 | ahcy | -0.050 |
| ppp1r14ba | 0.159 | si:ch211-156b7.4 | -0.050 |
| isl2b | 0.158 | si:dkey-261m9.17 | -0.050 |
| id4 | 0.157 | baz1a | -0.050 |
| zgc:65894 | 0.156 | tuba8l | -0.049 |
| zgc:158291 | 0.156 | rrm2 | -0.049 |
| rbpms2a | 0.153 | msna | -0.049 |
| xpr1a | 0.153 | rpa3 | -0.048 |
| maptb | 0.152 | cks1b | -0.048 |
| gng2 | 0.152 | mibp | -0.048 |
| klf7b | 0.152 | banf1 | -0.048 |
| kif3cb | 0.152 | zgc:110540 | -0.048 |
| dpysl2b | 0.151 | fen1 | -0.048 |
| zfhx3 | 0.151 | CABZ01005379.1 | -0.047 |
| rtn1a | 0.149 | neurod4 | -0.047 |
| cd99l2 | 0.145 | zgc:110216 | -0.047 |
| cxxc4 | 0.144 | zgc:165555.3 | -0.046 |