microtubule-associated protein 6b
ZFIN 
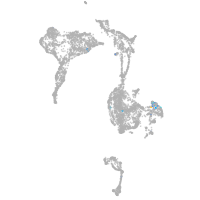
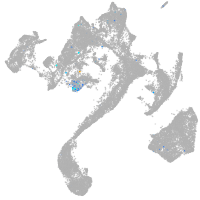
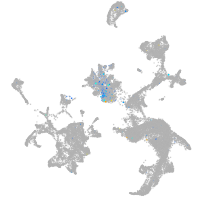
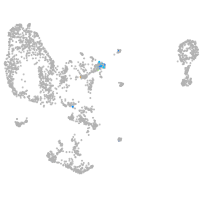
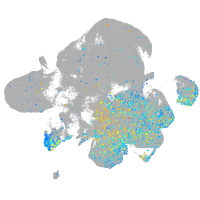
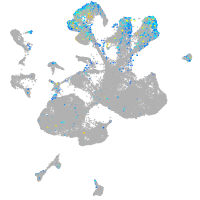
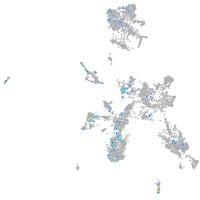
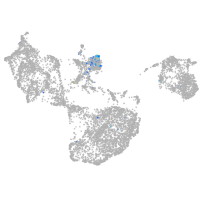
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| eno2 | 0.297 | hmgb2a | -0.188 |
| rtn1b | 0.294 | mdka | -0.102 |
| stmn2a | 0.290 | ahcy | -0.100 |
| ywhag2 | 0.283 | hmga1a | -0.099 |
| zgc:65894 | 0.271 | stmn1a | -0.099 |
| rbpms2a | 0.270 | pcna | -0.098 |
| zgc:153426 | 0.264 | fabp7a | -0.096 |
| stxbp1a | 0.262 | dut | -0.094 |
| gng3 | 0.256 | rrm1 | -0.094 |
| stx1b | 0.254 | msi1 | -0.094 |
| islr2 | 0.251 | lbr | -0.091 |
| syt2a | 0.246 | nutf2l | -0.091 |
| inab | 0.245 | ccnd1 | -0.089 |
| cplx2l | 0.245 | mki67 | -0.087 |
| elavl3 | 0.244 | chaf1a | -0.087 |
| snap25a | 0.244 | tuba8l | -0.085 |
| sncb | 0.240 | mcm7 | -0.085 |
| ppp1r14ba | 0.238 | msna | -0.084 |
| maptb | 0.235 | her15.1 | -0.084 |
| sprn | 0.234 | ccna2 | -0.083 |
| sv2a | 0.233 | eef1da | -0.083 |
| elavl4 | 0.232 | selenoh | -0.082 |
| tuba2 | 0.232 | hmgb2b | -0.080 |
| id4 | 0.229 | cks1b | -0.079 |
| isl2b | 0.227 | dek | -0.078 |
| tkta | 0.227 | COX7A2 (1 of many) | -0.078 |
| gabrb4 | 0.227 | rrm2 | -0.078 |
| mllt11 | 0.227 | si:ch211-156b7.4 | -0.076 |
| rbfox1 | 0.225 | rpa3 | -0.075 |
| gap43 | 0.225 | tspan7 | -0.074 |
| xpr1a | 0.224 | banf1 | -0.074 |
| rbpms2b | 0.223 | neurod4 | -0.074 |
| stmn4l | 0.218 | mibp | -0.074 |
| st8sia5 | 0.217 | lig1 | -0.074 |
| syt1a | 0.216 | cx43.4 | -0.074 |