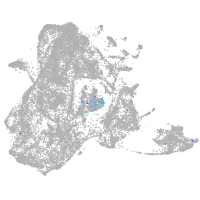
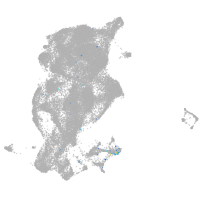
microtubule-associated protein 6a
ZFIN 
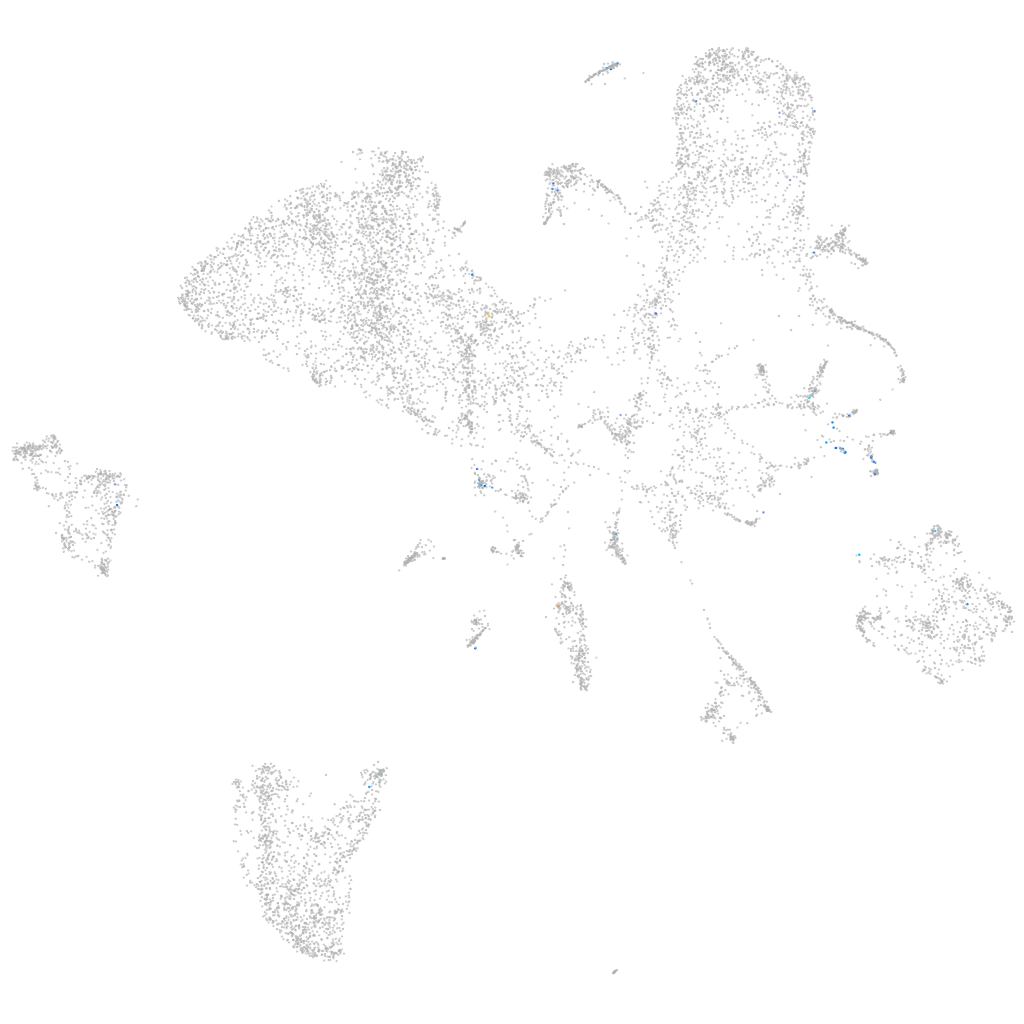
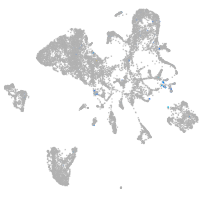
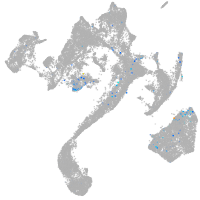
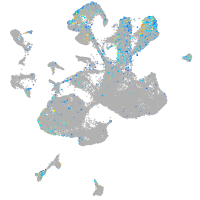
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-117c9.1 | 0.328 | aldob | -0.054 |
| BX664625.1 | 0.266 | ahcy | -0.049 |
| CU929145.2 | 0.240 | aldh7a1 | -0.045 |
| lingo4b | 0.234 | nupr1b | -0.044 |
| LOC100334937 | 0.215 | adka | -0.043 |
| CR589947.1 | 0.211 | rpl36a | -0.043 |
| mmp17a | 0.206 | mat1a | -0.041 |
| fgf11b | 0.205 | si:dkey-16p21.8 | -0.041 |
| gng3 | 0.200 | fbp1b | -0.041 |
| myo1hb | 0.188 | atp5f1c | -0.041 |
| sncb | 0.185 | sdr16c5b | -0.040 |
| AL645792.1 | 0.183 | shmt1 | -0.040 |
| LO018363.1 | 0.183 | ckba | -0.040 |
| aldoca | 0.182 | rps20 | -0.039 |
| ank3a | 0.180 | cx32.3 | -0.039 |
| nptna | 0.179 | hdlbpa | -0.039 |
| pou6f2 | 0.179 | eef1da | -0.038 |
| ptprn2 | 0.174 | tdo2a | -0.038 |
| ssh1b | 0.171 | upb1 | -0.037 |
| BX936305.2 | 0.167 | aldh1l1 | -0.037 |
| pitpnab | 0.167 | gapdh | -0.037 |
| setd1bb | 0.167 | eno3 | -0.037 |
| insm1a | 0.163 | cat | -0.037 |
| smarcd3a | 0.159 | ak2 | -0.037 |
| lmo1 | 0.157 | isoc2 | -0.037 |
| itm2ca | 0.155 | igfbp2a | -0.036 |
| ndufa4l2a | 0.154 | pgm1 | -0.036 |
| XLOC-008935 | 0.153 | cdo1 | -0.036 |
| kctd4 | 0.153 | si:ch73-281k2.5 | -0.036 |
| cadm2b | 0.152 | scp2a | -0.036 |
| ins | 0.151 | gcshb | -0.036 |
| coro2bb | 0.149 | ambp | -0.036 |
| neurod1 | 0.149 | mthfd1b | -0.036 |
| zmp:0000000619 | 0.148 | dap | -0.036 |
| si:dkey-153k10.9 | 0.146 | ttc36 | -0.035 |