mitogen-activated protein kinase kinase kinase 21
ZFIN 
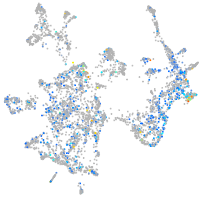
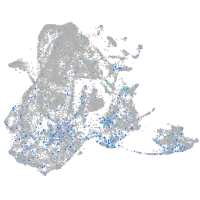
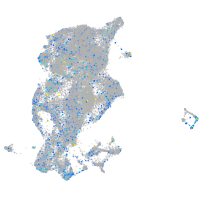
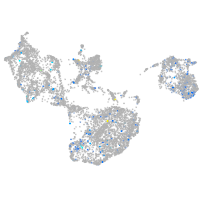
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| id1 | 0.166 | ckbb | -0.120 |
| cx43 | 0.156 | ptmab | -0.105 |
| wls | 0.151 | hmgn6 | -0.099 |
| zfp36l1a | 0.148 | snap25b | -0.099 |
| tspan36 | 0.147 | ptmaa | -0.098 |
| her9 | 0.144 | atp6v0cb | -0.097 |
| cldn5a | 0.142 | rnasekb | -0.091 |
| vamp3 | 0.140 | gapdhs | -0.090 |
| gas1b | 0.139 | stmn1b | -0.089 |
| her6 | 0.138 | gpm6aa | -0.084 |
| yap1 | 0.136 | ndrg4 | -0.084 |
| pmela | 0.134 | tuba1c | -0.082 |
| col4a6 | 0.129 | sypb | -0.081 |
| fabp11a | 0.128 | crx | -0.080 |
| dct | 0.128 | syt5b | -0.080 |
| cd63 | 0.128 | vamp2 | -0.080 |
| col18a1a | 0.128 | atpv0e2 | -0.079 |
| slc45a2 | 0.127 | atp1b2a | -0.079 |
| sparc | 0.127 | gpm6ab | -0.078 |
| adi1 | 0.127 | epb41a | -0.077 |
| rnaseka | 0.126 | rtn1a | -0.076 |
| zgc:158689 | 0.126 | atp1a3a | -0.074 |
| bace2 | 0.126 | scrt2 | -0.074 |
| crtap | 0.125 | ppp1r14c | -0.073 |
| eif4ebp3l | 0.125 | gng3 | -0.072 |
| slc38a5b | 0.124 | si:ch73-1a9.3 | -0.072 |
| zgc:110591 | 0.122 | rs1a | -0.071 |
| cdon | 0.122 | sncb | -0.070 |
| selenoh | 0.121 | pcbp3 | -0.070 |
| atp1b1a | 0.120 | neurod1 | -0.069 |
| stra6 | 0.120 | gng13b | -0.068 |
| tyrp1b | 0.120 | jpt1b | -0.068 |
| otx2a | 0.119 | mid1ip1b | -0.068 |
| pttg1ipb | 0.119 | golga7ba | -0.067 |
| col4a5 | 0.119 | pvalb2 | -0.067 |