"mannosidase, alpha, class 1B, member 1b"
ZFIN 
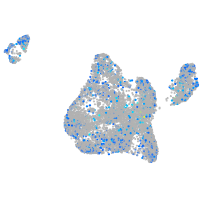
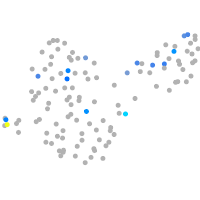
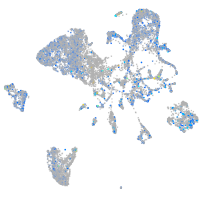
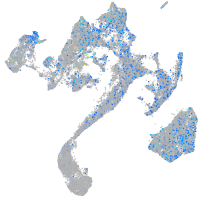
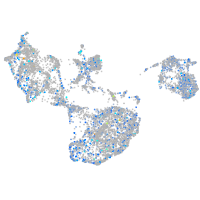
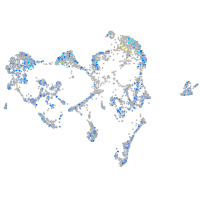
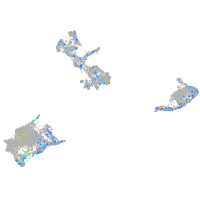
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-002522 | 0.185 | bms1 | -0.111 |
| rbm47 | 0.180 | apoc1 | -0.110 |
| hnf1ba | 0.176 | zmp:0000000624 | -0.104 |
| degs1 | 0.176 | snrnp70 | -0.102 |
| tspan35 | 0.175 | gnl3 | -0.102 |
| ezra | 0.175 | si:ch73-1a9.3 | -0.101 |
| zgc:56493 | 0.174 | ddx18 | -0.100 |
| cd164 | 0.173 | npm1a | -0.099 |
| si:ch73-368j24.14 | 0.171 | noc2l | -0.097 |
| tob1a | 0.169 | marcksb | -0.097 |
| itga6a | 0.169 | dkc1 | -0.096 |
| BX465848.2 | 0.169 | cdx4 | -0.095 |
| tegt | 0.167 | si:dkey-159f12.2 | -0.093 |
| bsg | 0.166 | apoeb | -0.093 |
| mt-cyb | 0.164 | marcksl1b | -0.092 |
| birc5b | 0.162 | wu:fb97g03 | -0.092 |
| mt-nd1 | 0.161 | snu13b | -0.091 |
| mt-nd4 | 0.160 | hspb1 | -0.090 |
| ch25hl3 | 0.159 | utp11l | -0.090 |
| fosab | 0.156 | tal1 | -0.090 |
| cdh17 | 0.156 | wdr46 | -0.089 |
| etnk1 | 0.156 | etv2 | -0.089 |
| mt-nd2 | 0.155 | tardbp | -0.088 |
| iqgap2 | 0.154 | COX7A2 | -0.088 |
| si:rp71-17i16.6 | 0.153 | ebna1bp2 | -0.087 |
| tpte | 0.153 | pprc1 | -0.086 |
| ccdc47 | 0.152 | ncl | -0.086 |
| sgk1 | 0.152 | si:ch211-113a14.18 | -0.086 |
| rnaseka | 0.151 | nhp2 | -0.085 |
| pgrmc1 | 0.151 | fbl | -0.085 |
| slc2a12 | 0.150 | rcc2 | -0.084 |
| bcam | 0.148 | nop2 | -0.083 |
| epcam | 0.147 | rrp1 | -0.083 |
| uqcrc1 | 0.147 | mphosph10 | -0.082 |
| mt-co1 | 0.146 | rrp15 | -0.081 |