v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b)
ZFIN 
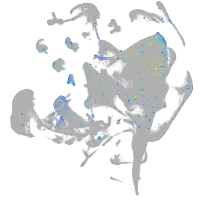
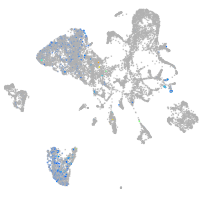
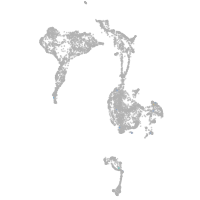
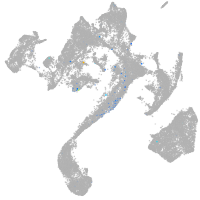
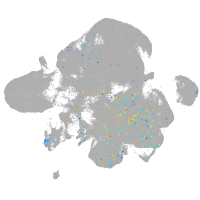
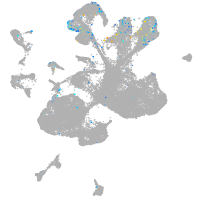
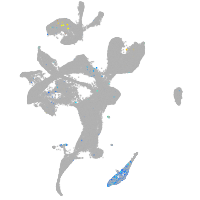
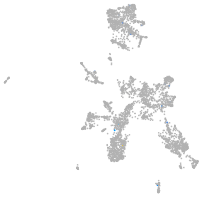
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU914144.1 | 0.306 | msna | -0.044 |
| rbfox1 | 0.129 | mki67 | -0.043 |
| cplx2l | 0.123 | banf1 | -0.042 |
| rbfox3a | 0.115 | ccnd1 | -0.041 |
| snap25a | 0.114 | vamp3 | -0.041 |
| elavl4 | 0.104 | chaf1a | -0.040 |
| gng3 | 0.103 | pcna | -0.040 |
| stmn2a | 0.102 | ccna2 | -0.039 |
| tubb2 | 0.097 | lig1 | -0.039 |
| pbx3b | 0.096 | akap12b | -0.038 |
| rtn1b | 0.095 | anp32e | -0.038 |
| stxbp1a | 0.095 | tpx2 | -0.038 |
| stx1b | 0.092 | ccnb1 | -0.037 |
| ywhag2 | 0.092 | cx43.4 | -0.037 |
| tmsb2 | 0.091 | dek | -0.037 |
| nefmb | 0.090 | tspan7 | -0.037 |
| cdk5r1b | 0.088 | ube2c | -0.037 |
| tmem59l | 0.088 | anp32b | -0.036 |
| sncb | 0.087 | dlgap5 | -0.036 |
| syt2a | 0.087 | hspb1 | -0.036 |
| gap43 | 0.086 | id1 | -0.036 |
| stmn1b | 0.085 | lbr | -0.036 |
| zgc:65894 | 0.085 | mad2l1 | -0.036 |
| id4 | 0.084 | si:ch211-286b5.5 | -0.036 |
| inab | 0.084 | sox19a | -0.036 |
| map1aa | 0.084 | zgc:110425 | -0.036 |
| dpysl3 | 0.082 | fbxo5 | -0.035 |
| gjd1a | 0.082 | rrm1 | -0.035 |
| mllt11 | 0.082 | cdca8 | -0.034 |
| mtus2a | 0.082 | mibp | -0.034 |
| kcnc3a | 0.082 | smc4 | -0.034 |
| map1b | 0.081 | aldob | -0.033 |
| tuba1c | 0.081 | aurkb | -0.033 |
| tuba2 | 0.081 | cdca7a | -0.033 |
| calm1a | 0.081 | cdca7b | -0.033 |