lysosomal trafficking regulator
ZFIN 
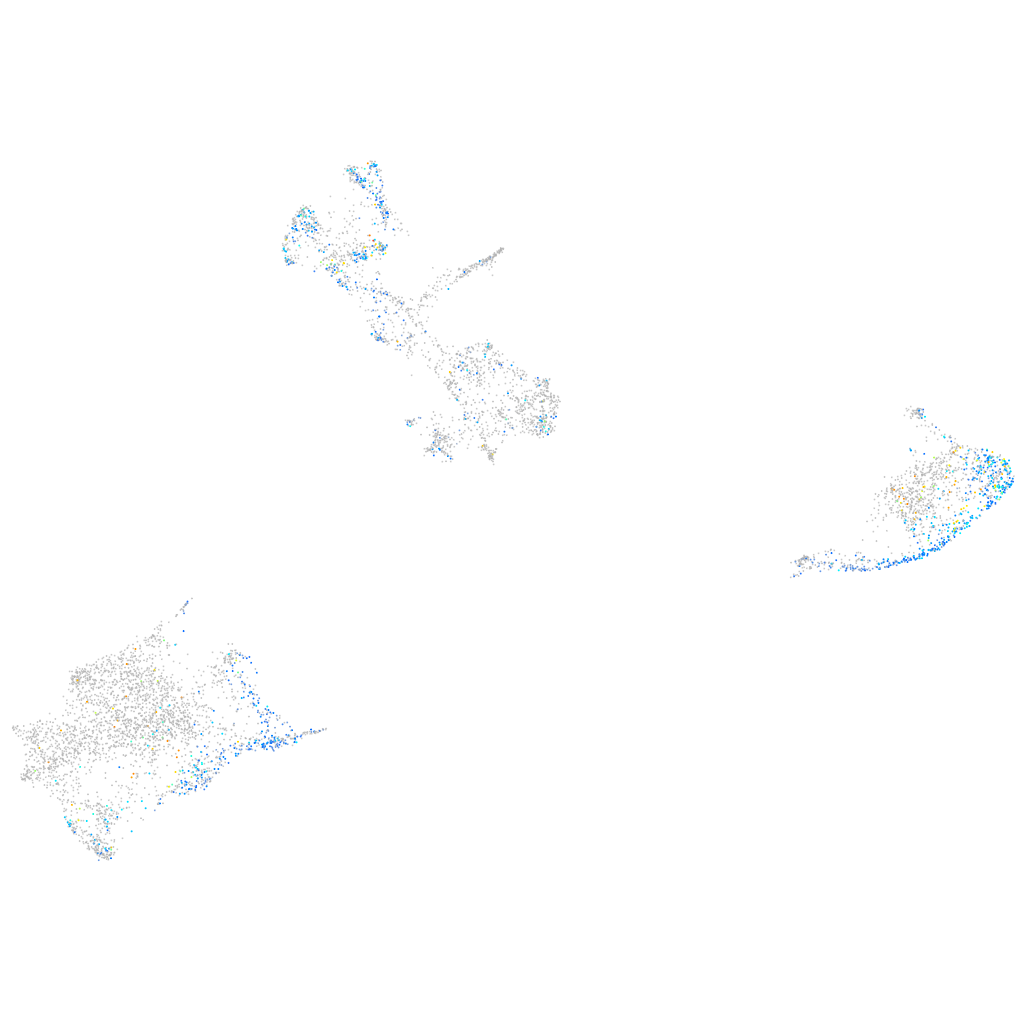
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.300 | CABZ01021592.1 | -0.169 |
| kita | 0.288 | si:dkey-251i10.2 | -0.150 |
| oca2 | 0.268 | uraha | -0.129 |
| slc39a10 | 0.262 | si:ch211-222l21.1 | -0.118 |
| atp11a | 0.258 | hbbe1.3 | -0.100 |
| slc45a2 | 0.254 | si:ch73-1a9.3 | -0.099 |
| slc24a5 | 0.250 | hmgn2 | -0.096 |
| slc29a3 | 0.243 | tmem130 | -0.095 |
| lamp1a | 0.242 | paics | -0.095 |
| kcnj13 | 0.241 | hbae3 | -0.092 |
| tyrp1a | 0.240 | ptmaa | -0.091 |
| prkar1b | 0.240 | si:ch211-251b21.1 | -0.090 |
| tyr | 0.239 | tuba1c | -0.085 |
| pmela | 0.238 | ptmab | -0.083 |
| slc7a5 | 0.235 | marcksl1a | -0.082 |
| dct | 0.231 | prdx5 | -0.081 |
| atp6v0ca | 0.230 | aldob | -0.079 |
| mchr2 | 0.228 | h3f3a | -0.079 |
| mtbl | 0.228 | hbae1.1 | -0.078 |
| tyrp1b | 0.227 | hmgb1b | -0.078 |
| atp6v0a2b | 0.224 | gch2 | -0.078 |
| pah | 0.224 | zgc:153031 | -0.076 |
| si:zfos-943e10.1 | 0.221 | stmn1b | -0.076 |
| CDK18 | 0.220 | hbbe1.1 | -0.076 |
| slc24a4a | 0.218 | cyb5a | -0.075 |
| slc22a2 | 0.218 | mdh1aa | -0.074 |
| opn5 | 0.216 | tmsb | -0.073 |
| birc7 | 0.216 | si:ch73-281n10.2 | -0.072 |
| sypl1 | 0.214 | hmgb3a | -0.071 |
| slc29a1a | 0.214 | gpm6aa | -0.071 |
| mc1r | 0.213 | aqp3a | -0.071 |
| bace2 | 0.213 | hmgb2b | -0.071 |
| spra | 0.212 | fabp7a | -0.070 |
| si:ch211-195b13.1 | 0.212 | elavl3 | -0.070 |
| myo5aa | 0.211 | stmn1a | -0.070 |