lysophospholipase 2
ZFIN 
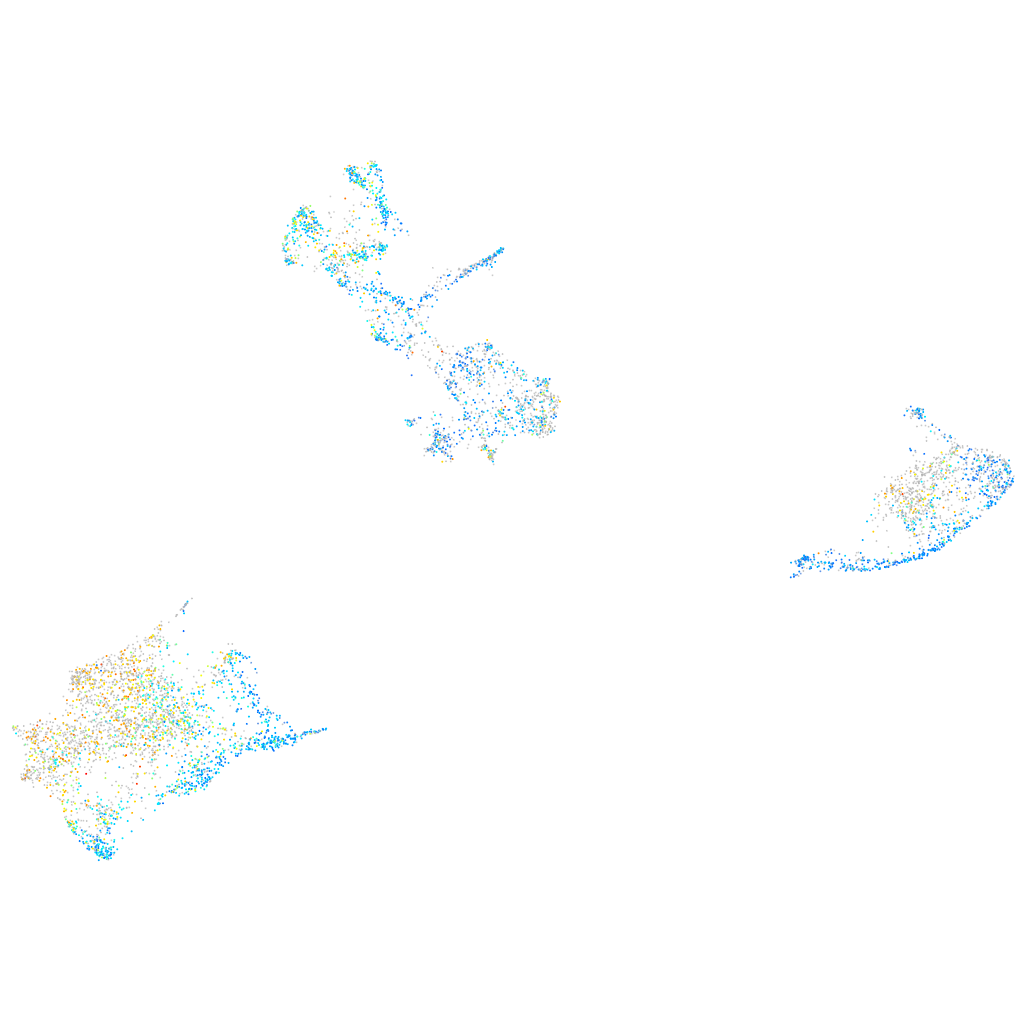
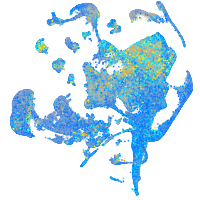
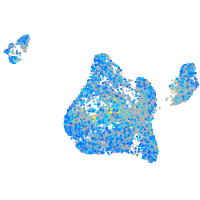
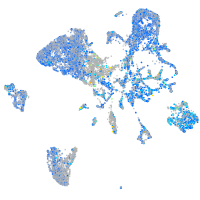
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gmps | 0.229 | tyrp1b | -0.130 |
| slc2a15a | 0.213 | pmela | -0.121 |
| impdh1b | 0.212 | zgc:91968 | -0.120 |
| prps1a | 0.187 | tyrp1a | -0.108 |
| glulb | 0.182 | anxa1a | -0.108 |
| atic | 0.182 | dct | -0.105 |
| paics | 0.181 | icn | -0.104 |
| pltp | 0.173 | nova2 | -0.099 |
| ivns1abpa | 0.169 | oca2 | -0.096 |
| mdh1aa | 0.167 | si:ch211-222l21.1 | -0.092 |
| ppat | 0.166 | ptmaa | -0.087 |
| gart | 0.163 | mtbl | -0.086 |
| psph | 0.162 | tuba1a | -0.082 |
| phyhd1 | 0.161 | s100a10b | -0.082 |
| adsl | 0.160 | prkar1b | -0.081 |
| zgc:110789 | 0.160 | foxp4 | -0.078 |
| shmt1 | 0.158 | tuba1c | -0.077 |
| akap12a | 0.155 | elavl3 | -0.076 |
| pgm2 | 0.154 | kita | -0.073 |
| tagln3b | 0.153 | gpm6aa | -0.073 |
| pnp4a | 0.152 | tubb5 | -0.073 |
| pax7a | 0.152 | slc39a10 | -0.071 |
| phgdh | 0.150 | stmn1b | -0.070 |
| sdc2 | 0.150 | slc6a15 | -0.070 |
| fhl3a | 0.149 | marcksl1a | -0.069 |
| tpd52l1 | 0.148 | tmeff1b | -0.069 |
| tpd52 | 0.148 | ckbb | -0.068 |
| gpnmb | 0.147 | ptgr1 | -0.068 |
| psat1 | 0.143 | FO082781.1 | -0.068 |
| guk1a | 0.143 | snap25b | -0.068 |
| si:dkey-197i20.6 | 0.143 | si:ch73-389b16.1 | -0.068 |
| sytl2b | 0.143 | tox | -0.067 |
| inka1a | 0.142 | hsd20b2 | -0.065 |
| shmt2 | 0.142 | zdhhc2 | -0.065 |
| sod1 | 0.141 | nat8l | -0.065 |