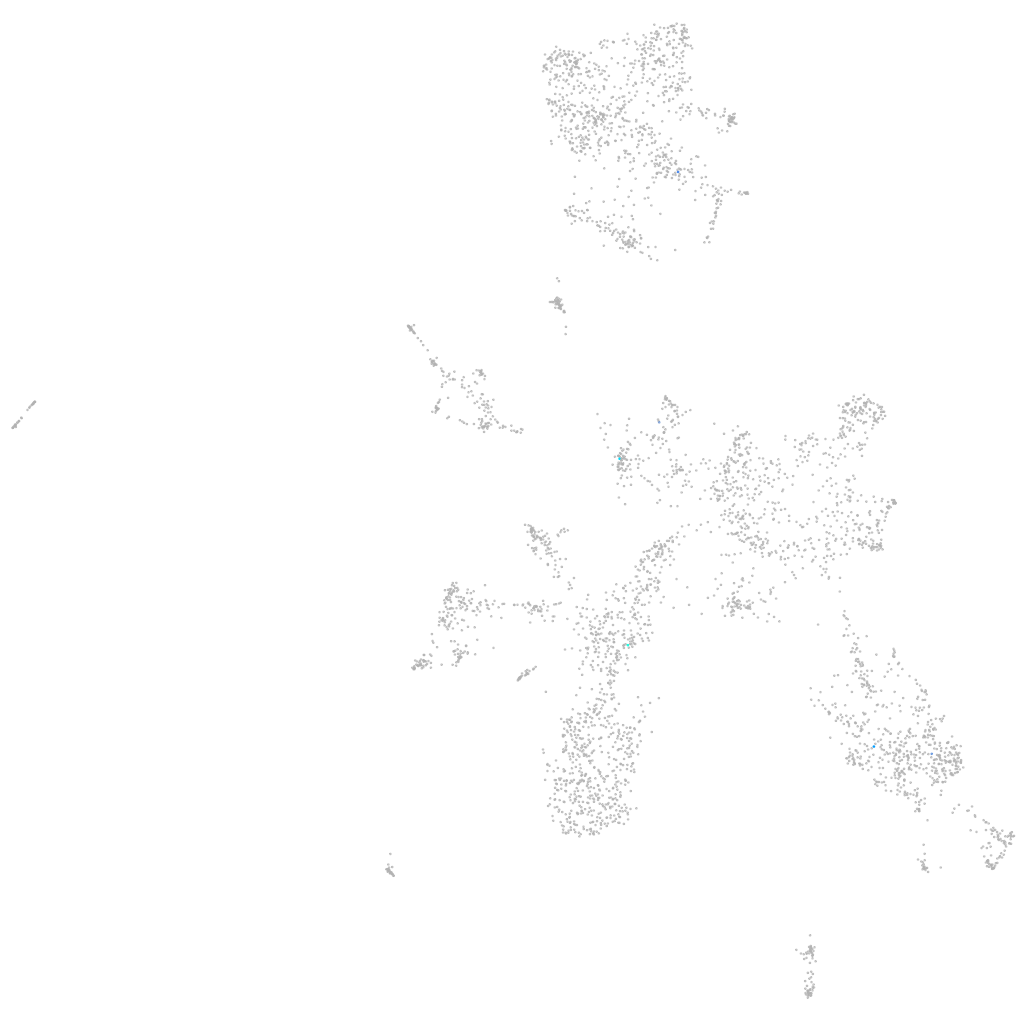
leucine rich repeat transmembrane neuronal 4 like 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-011609 | 0.587 | ndufb9 | -0.036 |
| trim55b | 0.444 | ndufa6 | -0.036 |
| si:ch211-238p8.24 | 0.418 | arf5 | -0.029 |
| tnni1b | 0.365 | rab2a | -0.029 |
| tal2 | 0.363 | atrx | -0.028 |
| XLOC-005983 | 0.335 | pfn2 | -0.028 |
| MFN1 | 0.328 | ndufa8 | -0.028 |
| CR751224.1 | 0.320 | sap30l | -0.028 |
| CABZ01084575.1 | 0.314 | snrnp27 | -0.028 |
| camkk1b | 0.310 | eif3ea | -0.027 |
| AGBL1 | 0.295 | nudc | -0.027 |
| CR751602.1 | 0.282 | ISCU (1 of many) | -0.027 |
| LOC101884250 | 0.279 | slc25a3b | -0.026 |
| LOC110438054 | 0.257 | romo1 | -0.026 |
| si:ch211-93g21.2 | 0.255 | zgc:92744 | -0.026 |
| dmbx1a | 0.248 | vps29 | -0.026 |
| taar12c | 0.242 | hdac1 | -0.026 |
| anks1ab | 0.240 | fkbp2 | -0.025 |
| CR376766.2 | 0.240 | sarnp | -0.025 |
| gad1a | 0.235 | tmed9 | -0.025 |
| abca4a | 0.232 | eif4a1b | -0.025 |
| LOC101883326 | 0.229 | canx | -0.025 |
| or109-1 | 0.226 | eif5a | -0.024 |
| slitrk3b | 0.220 | kdm1a | -0.024 |
| LOC108191536 | 0.217 | txndc9 | -0.024 |
| LOC100535347 | 0.216 | marcksb | -0.024 |
| grm8b | 0.213 | sf3b1 | -0.024 |
| kcna2a | 0.212 | ndufa2 | -0.024 |
| zgc:162972 | 0.208 | selenok | -0.024 |
| slitrk3a | 0.202 | fam133b | -0.024 |
| nphs1 | 0.200 | psap | -0.024 |
| LOC101883160 | 0.191 | csde1 | -0.024 |
| BX511265.2 | 0.182 | hmgn3 | -0.024 |
| rpp25b | 0.179 | sec61a1 | -0.024 |
| grin2aa | 0.174 | csnk2a1 | -0.024 |