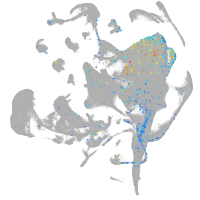
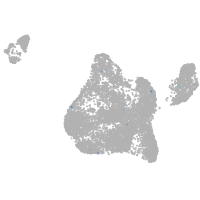
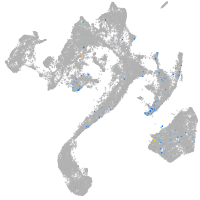
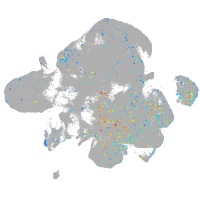
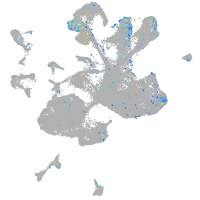
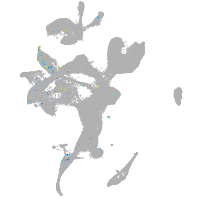
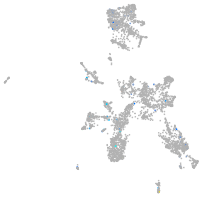
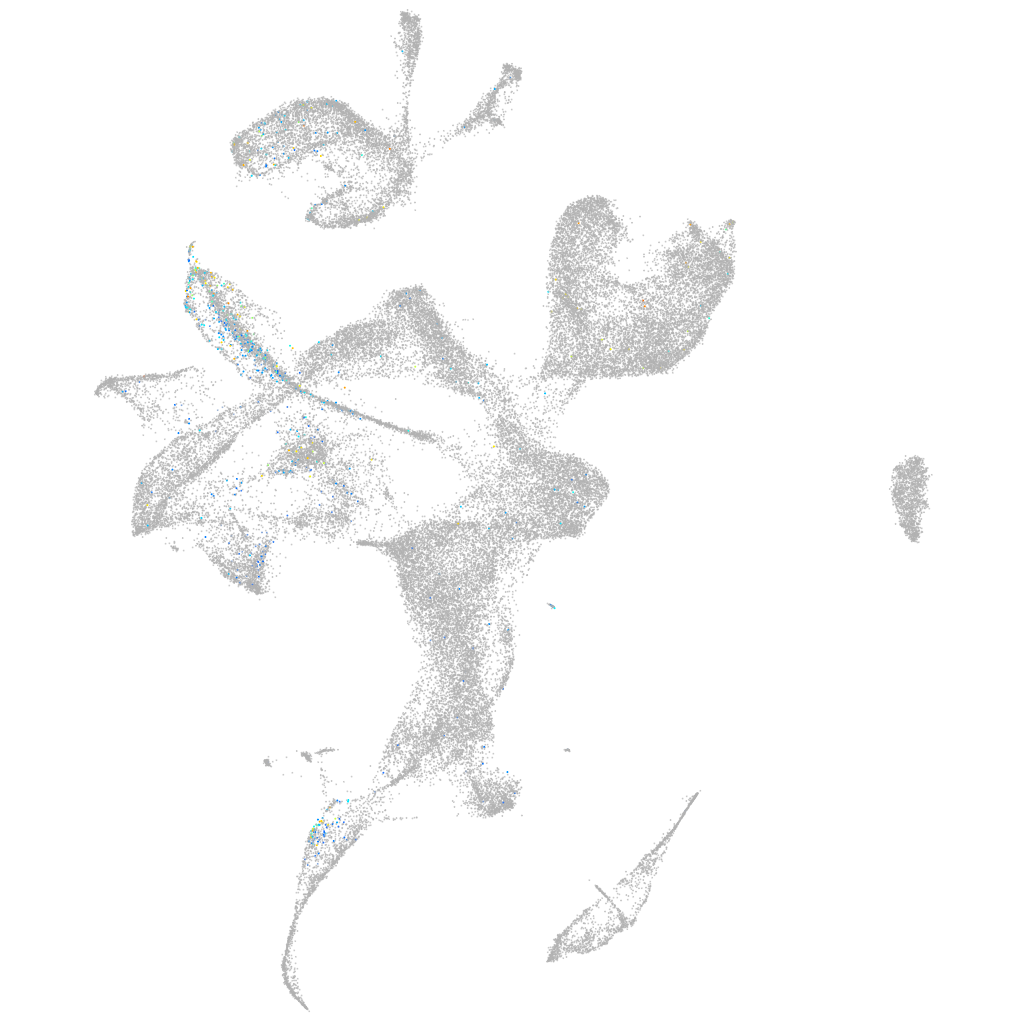leucine rich repeat neuronal 3a
ZFIN 
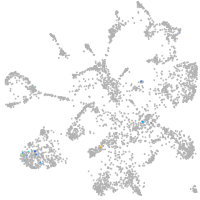
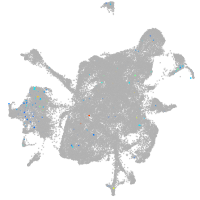
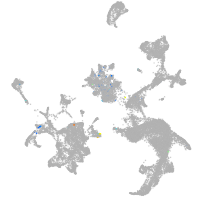
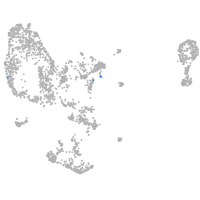
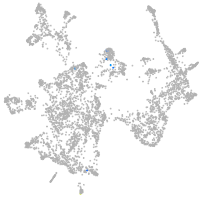
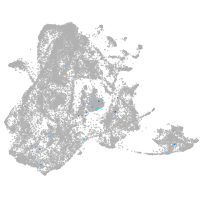
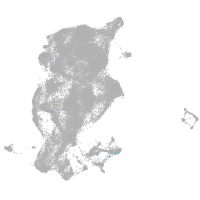
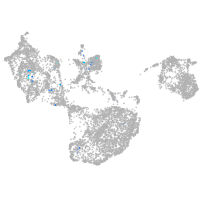
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rbpms2a | 0.167 | hmgb2a | -0.086 |
| rbpms2b | 0.165 | pcna | -0.046 |
| isl2b | 0.156 | fabp7a | -0.046 |
| inab | 0.156 | hmga1a | -0.045 |
| pou6f2 | 0.154 | stmn1a | -0.044 |
| islr2 | 0.141 | dut | -0.044 |
| FAM163B (1 of many) | 0.138 | nutf2l | -0.044 |
| xpr1a | 0.135 | mki67 | -0.044 |
| zgc:158291 | 0.132 | rrm1 | -0.043 |
| nrn1a | 0.131 | si:ch73-281n10.2 | -0.042 |
| chrnb3b | 0.126 | anp32b | -0.041 |
| zfhx3 | 0.122 | msi1 | -0.040 |
| atp2b3b | 0.122 | lbr | -0.040 |
| stmn2a | 0.122 | mdka | -0.040 |
| cplx2l | 0.120 | rpa3 | -0.039 |
| eno2 | 0.119 | cx43.4 | -0.039 |
| rtn1b | 0.118 | ccna2 | -0.039 |
| maptb | 0.117 | lig1 | -0.039 |
| chrna6 | 0.117 | neurod4 | -0.038 |
| cntn2 | 0.115 | hmgb2b | -0.038 |
| adcyap1b | 0.115 | zgc:110425 | -0.038 |
| adam11 | 0.115 | dek | -0.038 |
| syt2a | 0.114 | nasp | -0.038 |
| kcnc3b | 0.114 | her15.1 | -0.037 |
| shisal1a | 0.114 | ccnd1 | -0.037 |
| map7d2b | 0.113 | rrm2 | -0.037 |
| oaz2b | 0.112 | h2afvb | -0.037 |
| stmn4 | 0.112 | mcm6 | -0.037 |
| ptchd1 | 0.111 | mcm7 | -0.037 |
| gap43 | 0.111 | smc2 | -0.036 |
| BX247868.1 | 0.111 | CABZ01005379.1 | -0.036 |
| pou4f1 | 0.108 | chaf1a | -0.036 |
| gng3 | 0.107 | cks1b | -0.036 |
| elavl3 | 0.106 | COX7A2 (1 of many) | -0.036 |
| frmd3 | 0.106 | ran | -0.036 |