leucine rich repeat and coiled-coil centrosomal protein 1
ZFIN 
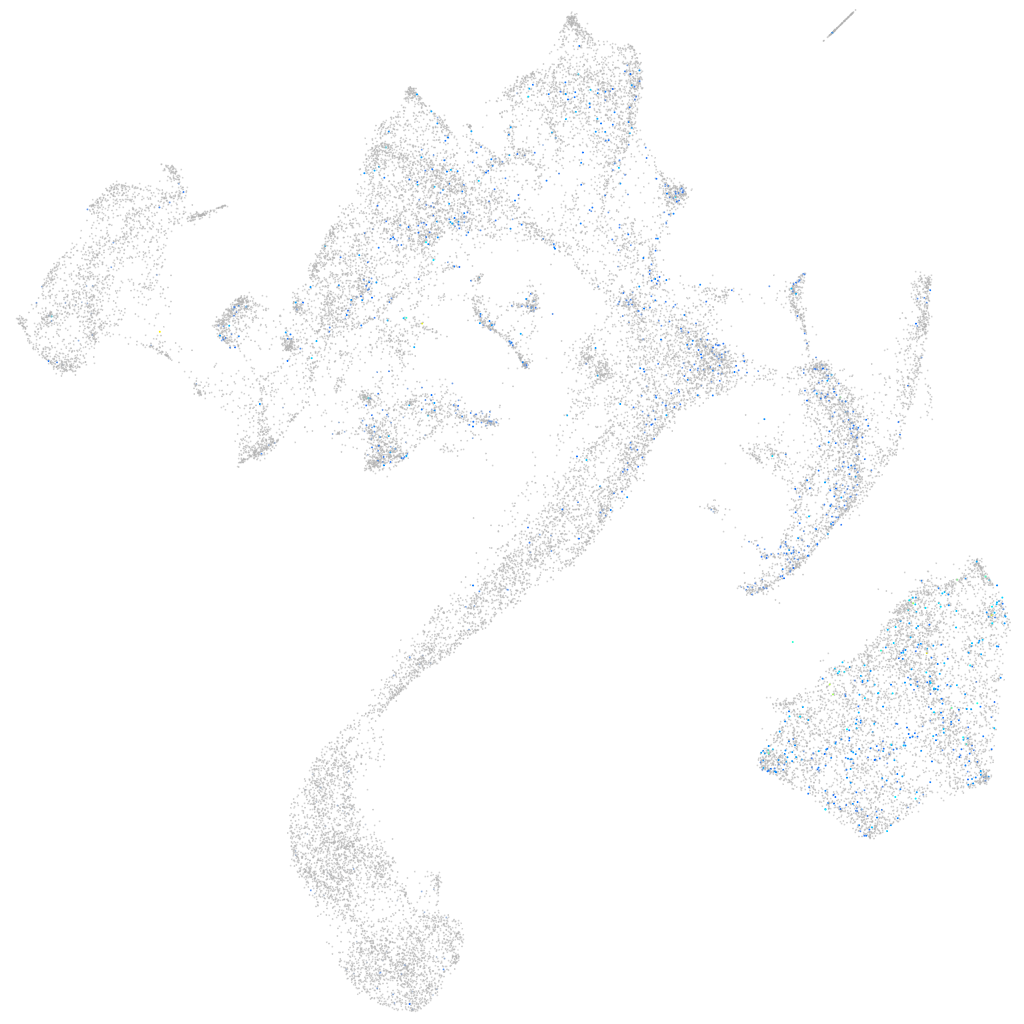
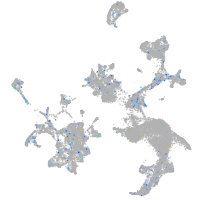
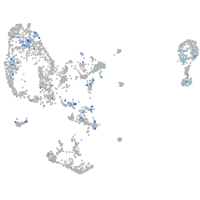
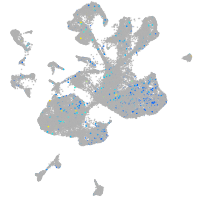
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mki67 | 0.155 | actc1b | -0.110 |
| tpx2 | 0.142 | ak1 | -0.097 |
| hmgb2a | 0.135 | aldoab | -0.097 |
| seta | 0.133 | atp2a1 | -0.095 |
| cenpf | 0.131 | ckma | -0.095 |
| top2a | 0.129 | rpl37 | -0.094 |
| aurkb | 0.128 | ttn.1 | -0.094 |
| nusap1 | 0.127 | ckmb | -0.094 |
| lbr | 0.126 | acta1b | -0.093 |
| mad2l1 | 0.126 | tmem38a | -0.093 |
| anp32b | 0.126 | ttn.2 | -0.093 |
| dek | 0.126 | tnnc2 | -0.093 |
| ptmab | 0.125 | atp5if1b | -0.090 |
| tubb2b | 0.125 | tpma | -0.090 |
| banf1 | 0.125 | zgc:114188 | -0.089 |
| ube2c | 0.125 | mybphb | -0.089 |
| anp32a | 0.124 | gamt | -0.089 |
| cdk1 | 0.123 | acta1a | -0.088 |
| si:ch211-222l21.1 | 0.123 | srl | -0.088 |
| plk1 | 0.123 | neb | -0.088 |
| ccnb1 | 0.122 | pabpc4 | -0.088 |
| smc1al | 0.121 | ldb3a | -0.088 |
| smc2 | 0.121 | mylpfa | -0.087 |
| lig1 | 0.121 | desma | -0.087 |
| zgc:110425 | 0.120 | CABZ01078594.1 | -0.086 |
| hmga1a | 0.119 | rps10 | -0.086 |
| si:ch211-288g17.3 | 0.119 | cav3 | -0.085 |
| hmgb2b | 0.118 | vdac3 | -0.084 |
| tuba8l4 | 0.118 | rps17 | -0.084 |
| fbxo5 | 0.118 | ldb3b | -0.084 |
| cx43.4 | 0.118 | gapdh | -0.084 |
| hnrnpa1b | 0.117 | myl1 | -0.083 |
| hnrnpub | 0.116 | tnnt3a | -0.082 |
| chaf1a | 0.116 | nme2b.2 | -0.082 |
| si:ch73-1a9.3 | 0.116 | eno3 | -0.082 |