leucine rich repeat containing 6
ZFIN 
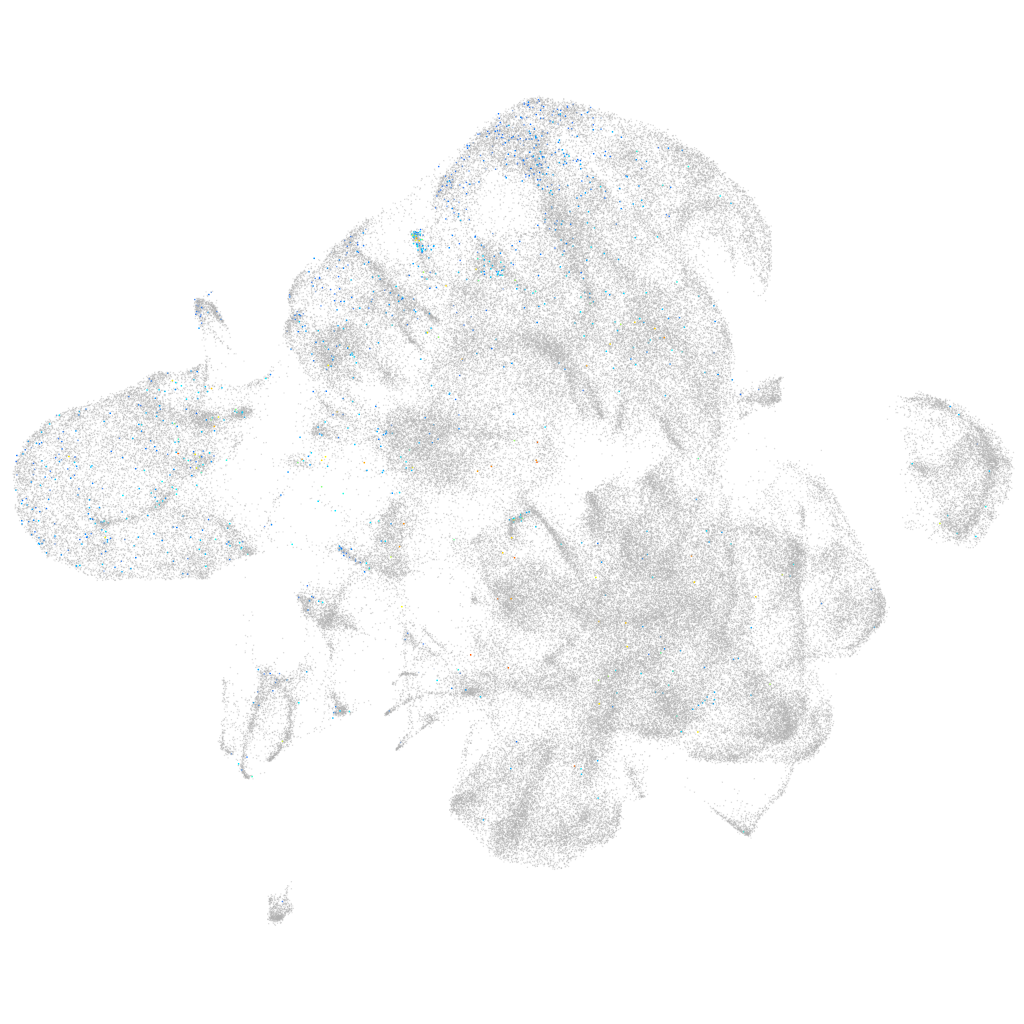
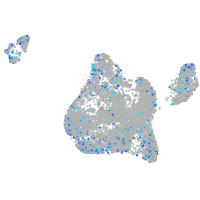
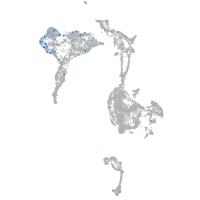
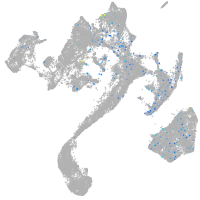
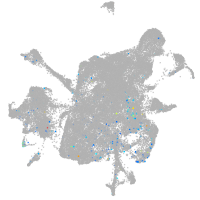
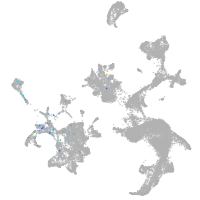
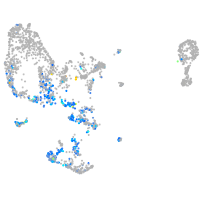
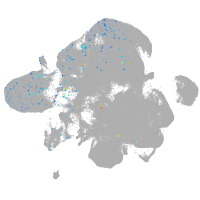
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccdc169 | 0.114 | stmn1b | -0.055 |
| daw1 | 0.108 | elavl3 | -0.053 |
| foxj1a | 0.100 | rtn1a | -0.050 |
| ccdc146 | 0.098 | ptmaa | -0.048 |
| tbata | 0.094 | tmsb | -0.046 |
| cfap157 | 0.090 | tuba1c | -0.044 |
| ccdc173 | 0.088 | gpm6ab | -0.042 |
| si:ch211-163l21.7 | 0.087 | gpm6aa | -0.041 |
| cfap52 | 0.086 | tubb5 | -0.039 |
| si:ch211-155m12.5 | 0.085 | ckbb | -0.039 |
| efcab1 | 0.082 | gng3 | -0.038 |
| dnaaf1 | 0.080 | COX3 | -0.037 |
| enkur | 0.078 | fez1 | -0.035 |
| bmp2a | 0.077 | mt-atp6 | -0.035 |
| si:dkey-27p23.3 | 0.076 | ywhah | -0.034 |
| RSPH1 | 0.074 | nova2 | -0.034 |
| si:ch211-248e11.2 | 0.074 | ppiab | -0.034 |
| ccdc65 | 0.073 | hmgb3a | -0.034 |
| armc3 | 0.072 | sncb | -0.034 |
| ak7a | 0.072 | rtn1b | -0.034 |
| cfap126 | 0.072 | elavl4 | -0.034 |
| ccdc114 | 0.071 | myt1b | -0.033 |
| hepacama | 0.070 | tmsb4x | -0.032 |
| dydc2 | 0.070 | stmn2a | -0.032 |
| si:ch211-226m7.4 | 0.069 | pvalb2 | -0.032 |
| crocc2 | 0.069 | mt-co2 | -0.032 |
| si:ch211-66e2.5 | 0.068 | atp6v0cb | -0.031 |
| si:dkey-42p14.3 | 0.067 | gap43 | -0.031 |
| fam183a | 0.067 | actc1b | -0.031 |
| spag6 | 0.066 | tmsb2 | -0.031 |
| CR932360.1 | 0.066 | celf2 | -0.031 |
| pacrg | 0.066 | stx1b | -0.031 |
| dnali1 | 0.065 | dpysl3 | -0.031 |
| fabp7b | 0.065 | pvalb1 | -0.031 |
| ropn1l | 0.064 | mt-nd1 | -0.030 |