leucine rich repeat containing 23
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis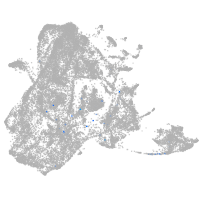 |
periderm |
fin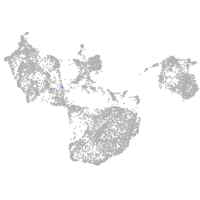 |
ionocytes / mucous |
pigment cells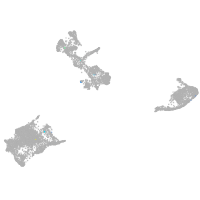 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| heg1 | 0.809 | ppiaa | -0.230 |
| col5a2b | 0.799 | csde1 | -0.223 |
| LOC100535347 | 0.799 | h3f3d | -0.216 |
| NC-002333.5 | 0.799 | serbp1a | -0.213 |
| BX294181.2 | 0.776 | chd4b | -0.211 |
| sytl5 | 0.774 | mfap1 | -0.196 |
| pard6ga | 0.764 | surf2 | -0.194 |
| armc9 | 0.741 | srsf5a | -0.193 |
| fras1 | 0.732 | cdca7b | -0.190 |
| umad1 | 0.729 | sf3b2 | -0.185 |
| zdhhc12b | 0.713 | si:ch73-281n10.2 | -0.181 |
| atp1a3a | 0.712 | psme3 | -0.179 |
| eif2b5 | 0.702 | cdk11b | -0.178 |
| si:ch211-261d7.6 | 0.691 | calm2a | -0.173 |
| CU655845.1 | 0.679 | anp32a | -0.171 |
| trim107 | 0.672 | hp1bp3 | -0.170 |
| brf2 | 0.667 | zmat2 | -0.168 |
| spice1 | 0.664 | tfdp1a | -0.166 |
| lhpp | 0.664 | smarca5 | -0.166 |
| si:ch211-214b16.2 | 0.655 | lrrc59 | -0.166 |
| si:dkeyp-59c12.1 | 0.650 | dido1 | -0.165 |
| FQ790211.1 | 0.645 | zc3h13 | -0.165 |
| HUS1B | 0.645 | tgif1 | -0.165 |
| wdyhv1 | 0.645 | dhx16 | -0.164 |
| trmt12 | 0.644 | pfn2l | -0.163 |
| si:dkey-174i8.1 | 0.640 | dnttip2 | -0.163 |
| rabgap1 | 0.639 | eif1axb | -0.163 |
| l3hypdh | 0.629 | htatsf1 | -0.163 |
| si:dkey-20i20.9 | 0.622 | ssrp1a | -0.163 |
| zgc:112998 | 0.619 | hmgb1a | -0.163 |
| pbrm1 | 0.616 | tbx16 | -0.162 |
| znf45l | 0.615 | setb | -0.161 |
| pimr67 | 0.605 | rbm4.3 | -0.161 |
| osbpl11 | 0.600 | ammecr1 | -0.161 |
| map3k5 | 0.595 | gps1 | -0.160 |