low density lipoprotein receptor-related protein 2a
ZFIN 
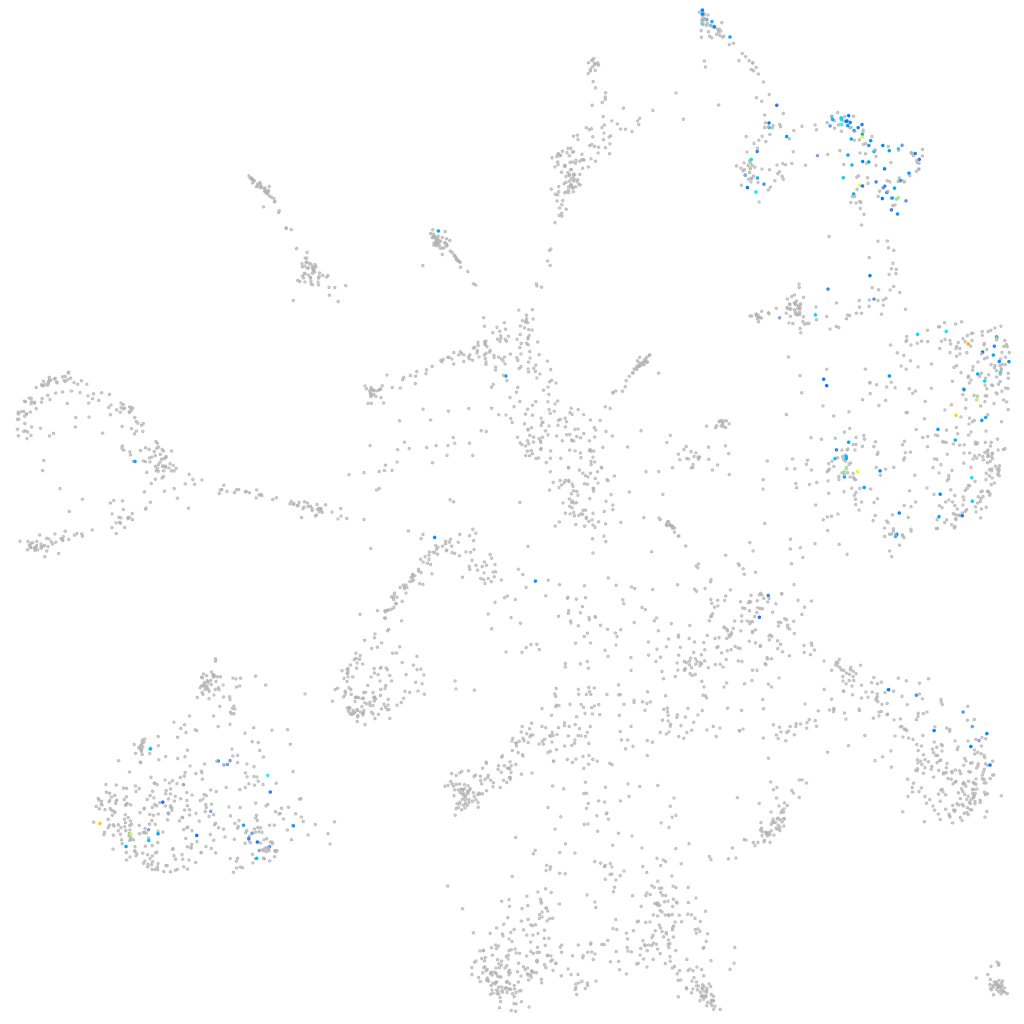
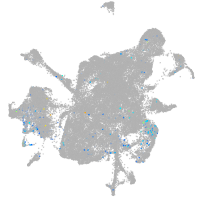
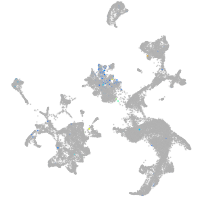
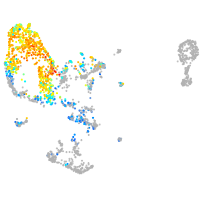
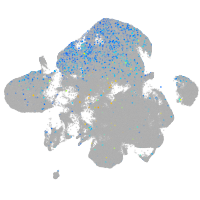
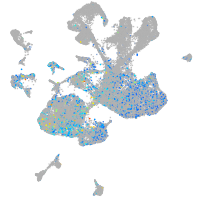
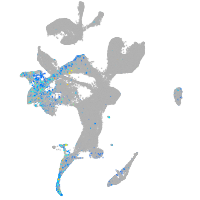
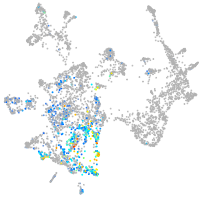
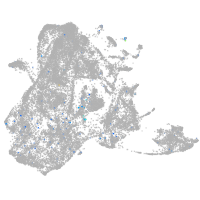
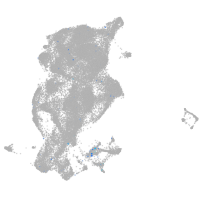
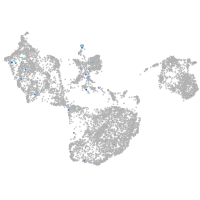
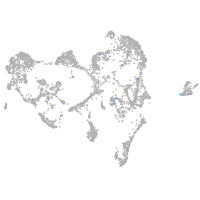
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.289 | BX088707.3 | -0.169 |
| mmel1 | 0.275 | myl9a | -0.135 |
| sfrp5 | 0.266 | tpm1 | -0.133 |
| jam2b | 0.263 | acta2 | -0.132 |
| lbx2 | 0.258 | tagln | -0.125 |
| alpi.1 | 0.253 | ptmaa | -0.112 |
| synpo2lb | 0.252 | myh11a | -0.111 |
| gstm.3 | 0.249 | mylkb | -0.106 |
| cxadr | 0.244 | foxf1 | -0.101 |
| tmem88b | 0.242 | lmod1b | -0.100 |
| bnc2 | 0.241 | gapdhs | -0.094 |
| lmod2a | 0.240 | tpm2 | -0.092 |
| bcam | 0.232 | myl6 | -0.091 |
| XLOC-015407 | 0.230 | fhl2a | -0.087 |
| XLOC-042222 | 0.228 | inka1a | -0.086 |
| krt94 | 0.228 | tmem119b | -0.085 |
| slc29a1b | 0.223 | XLOC-025423 | -0.084 |
| fabp11a | 0.222 | vdac1 | -0.084 |
| crb2a | 0.220 | ppiab | -0.081 |
| eppk1 | 0.219 | nkx2.3 | -0.081 |
| gata6 | 0.216 | myocd | -0.081 |
| LOC103910903 | 0.216 | XLOC-041870 | -0.079 |
| krt8 | 0.215 | sat1a.2 | -0.079 |
| gata5 | 0.212 | cald1b | -0.079 |
| alx4a | 0.204 | mylka | -0.079 |
| bmper | 0.203 | csrp1b | -0.078 |
| tnni1b | 0.202 | ntn5 | -0.078 |
| zfp36l1a | 0.201 | cnn1b | -0.078 |
| tgm2b | 0.198 | mcamb | -0.077 |
| epb41l5 | 0.198 | calm2a | -0.077 |
| mmp11a | 0.196 | desmb | -0.076 |
| smoc1 | 0.194 | si:ch211-62a1.3 | -0.074 |
| wt1b | 0.192 | rbpms2a | -0.074 |
| cd151 | 0.190 | gpia | -0.073 |
| angptl6 | 0.190 | pdgfrb | -0.072 |