LIM domain only 2 (rhombotin-like 1)
ZFIN 
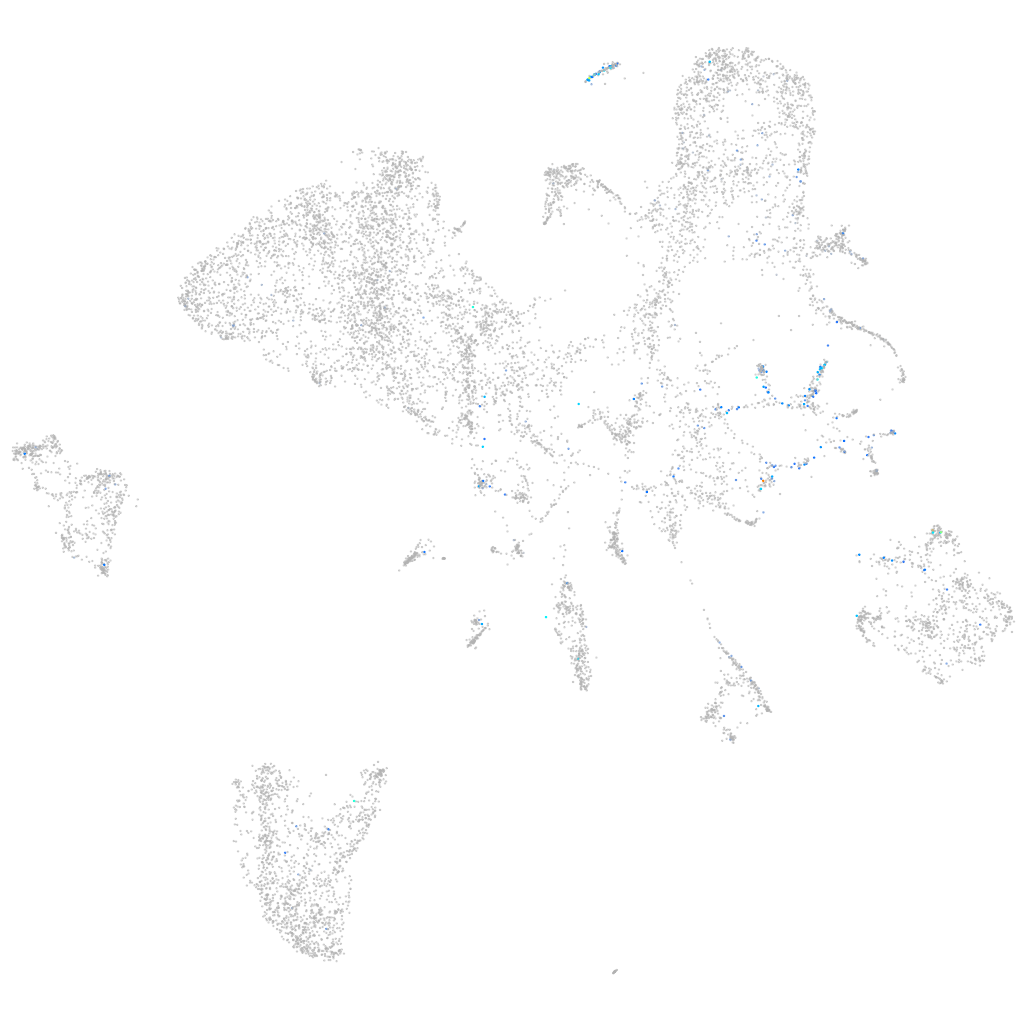
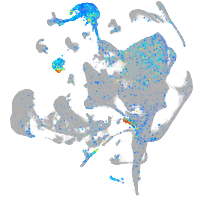
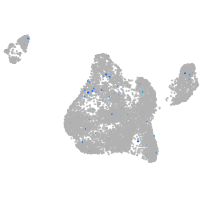
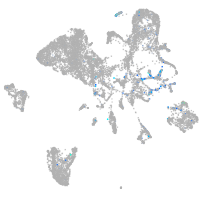
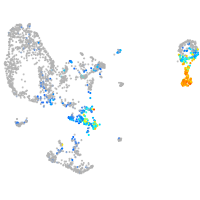
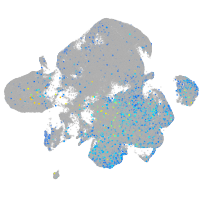
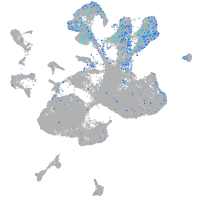
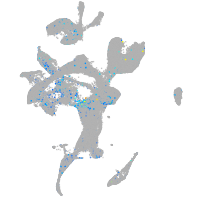
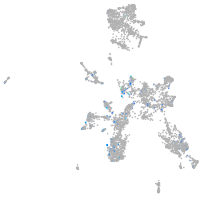
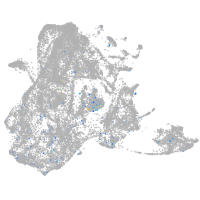
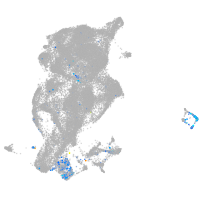
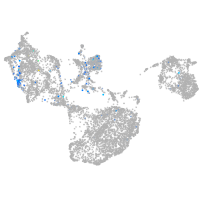
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.250 | ahcy | -0.109 |
| etv2 | 0.246 | gapdh | -0.107 |
| tal1 | 0.231 | gamt | -0.099 |
| egr4 | 0.228 | gatm | -0.093 |
| dll4 | 0.226 | mat1a | -0.093 |
| insm1b | 0.212 | aldob | -0.092 |
| adcyap1a | 0.206 | aldh6a1 | -0.088 |
| fev | 0.206 | cx32.3 | -0.088 |
| trpa1b | 0.205 | fabp3 | -0.088 |
| mir375-2 | 0.197 | bhmt | -0.086 |
| si:ch211-261a10.5 | 0.197 | fbp1b | -0.085 |
| neurod1 | 0.196 | aldh7a1 | -0.085 |
| ret | 0.184 | gnmt | -0.083 |
| zgc:101731 | 0.182 | aqp12 | -0.082 |
| XLOC-007078 | 0.181 | gstt1a | -0.081 |
| rgs16 | 0.178 | apoa1b | -0.081 |
| slc35g2a | 0.175 | nupr1b | -0.081 |
| tspan7b | 0.172 | hdlbpa | -0.080 |
| calm1b | 0.170 | apoa4b.1 | -0.080 |
| nmu | 0.169 | agxtb | -0.080 |
| ccka | 0.167 | dap | -0.079 |
| c2cd4a | 0.167 | glud1b | -0.078 |
| lmx1ba | 0.167 | agxta | -0.077 |
| pax6b | 0.166 | mgst1.2 | -0.076 |
| hepacam2 | 0.166 | scp2a | -0.075 |
| mir7a-1 | 0.165 | apoc2 | -0.075 |
| vamp2 | 0.162 | eno3 | -0.074 |
| fli1a | 0.161 | ttc36 | -0.074 |
| myct1a | 0.161 | afp4 | -0.074 |
| elovl1a | 0.160 | nipsnap3a | -0.074 |
| pcsk1nl | 0.159 | apoa2 | -0.074 |
| scgn | 0.158 | hspd1 | -0.073 |
| vat1 | 0.158 | LOC110437731 | -0.072 |
| slc7a14a | 0.158 | serpinf2b | -0.072 |
| sprn2 | 0.156 | ndrg2 | -0.072 |