limb and CNS expressed 1 like
ZFIN 
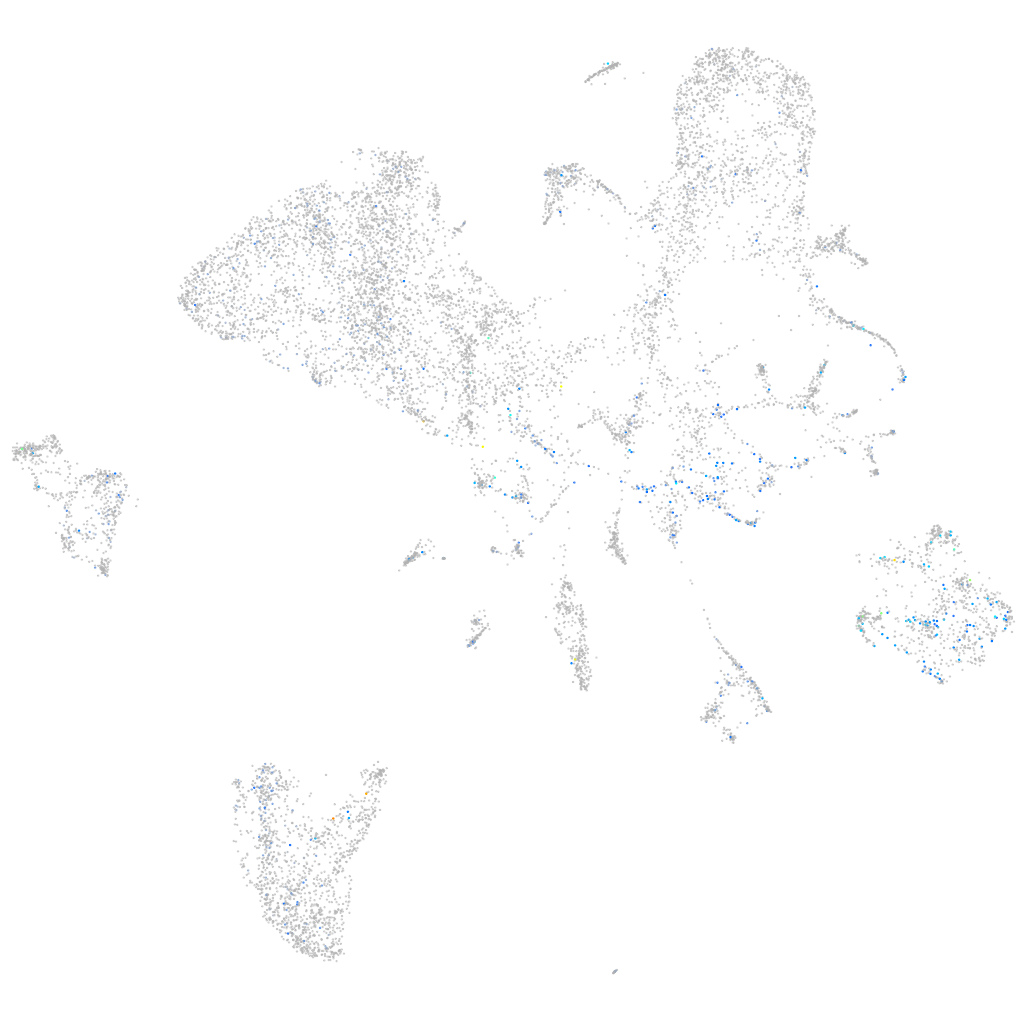
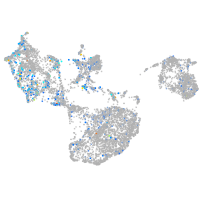
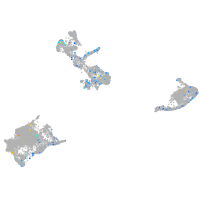
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| RF00069 | 0.172 | gapdh | -0.113 |
| marcksb | 0.159 | eno3 | -0.113 |
| akap12b | 0.155 | rpl37 | -0.107 |
| marcksl1b | 0.154 | ahcy | -0.104 |
| LOC101882264 | 0.152 | gamt | -0.101 |
| XLOC-016343 | 0.152 | sod1 | -0.099 |
| hmgb3a | 0.150 | atp5if1b | -0.097 |
| parp6a | 0.148 | rps10 | -0.096 |
| cx43.4 | 0.147 | prdx2 | -0.095 |
| si:ch211-152c2.3 | 0.145 | fbp1b | -0.094 |
| hmgb1b | 0.142 | suclg1 | -0.094 |
| anp32e | 0.142 | zgc:114188 | -0.092 |
| hspb1 | 0.141 | gstt1a | -0.092 |
| cdh6 | 0.140 | atp5l | -0.092 |
| hnrnpa0a | 0.140 | gpx4a | -0.091 |
| hnrnpa0b | 0.139 | lgals2b | -0.091 |
| h3f3d | 0.138 | aldob | -0.091 |
| hnrnpaba | 0.138 | mdh1aa | -0.091 |
| ewsr1a | 0.137 | apoc2 | -0.091 |
| seta | 0.136 | nme2b.1 | -0.091 |
| hmga1a | 0.136 | apoa4b.1 | -0.090 |
| syncrip | 0.136 | eef1da | -0.089 |
| ppig | 0.135 | gatm | -0.088 |
| si:ch211-137a8.4 | 0.135 | rps17 | -0.087 |
| khdrbs1a | 0.135 | scp2a | -0.086 |
| hmgn6 | 0.134 | sult2st2 | -0.086 |
| nono | 0.133 | pgk1 | -0.085 |
| hmgb2a | 0.133 | gstp1 | -0.085 |
| anp32b | 0.131 | pgam1a | -0.085 |
| acin1a | 0.131 | tpi1b | -0.084 |
| rsf1b.1 | 0.131 | fabp2 | -0.084 |
| nucks1a | 0.131 | ugt1a7 | -0.083 |
| lima1a | 0.130 | gsta.1 | -0.083 |
| si:ch73-1a9.3 | 0.130 | pklr | -0.083 |
| cbx3a | 0.130 | glud1b | -0.082 |