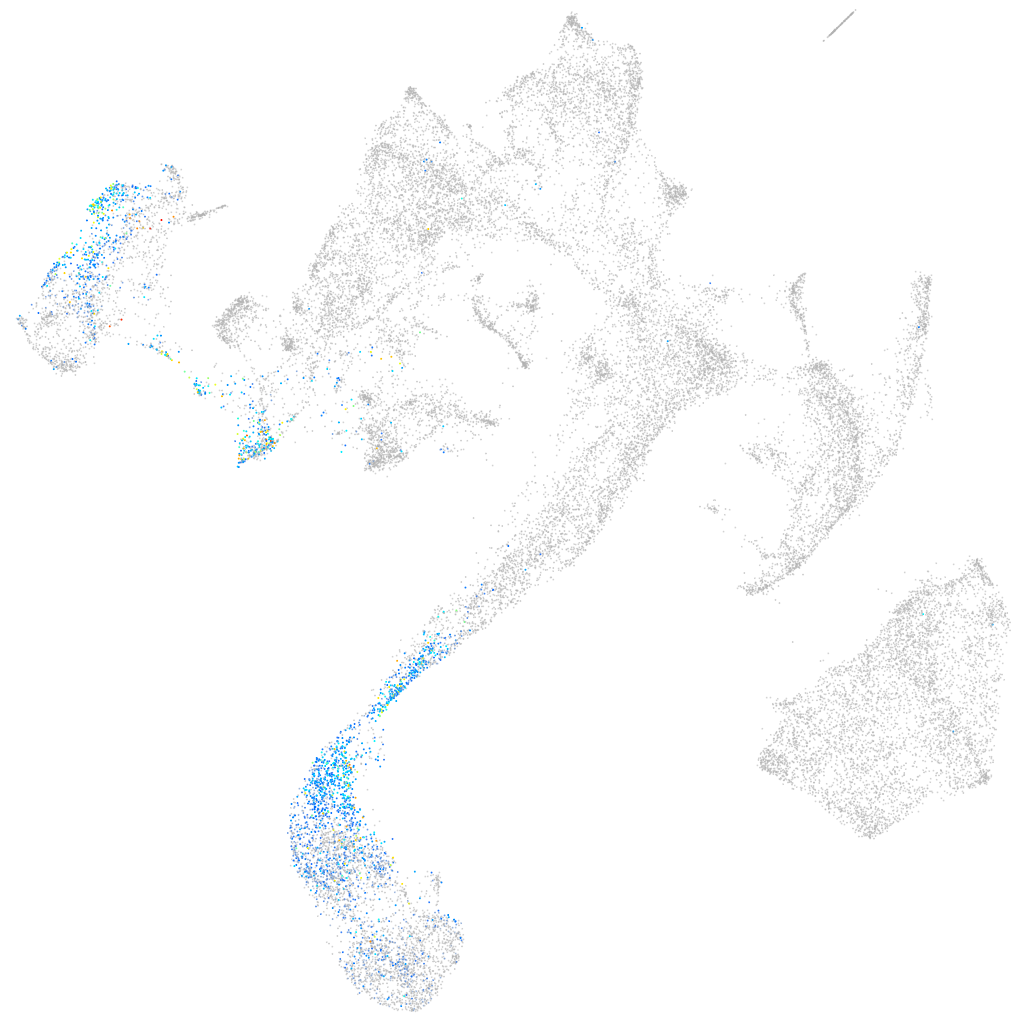
LIM domains containing 1b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cavin4b | 0.513 | hsp90ab1 | -0.389 |
| cavin4a | 0.500 | pabpc1a | -0.350 |
| zgc:158296 | 0.483 | h3f3d | -0.339 |
| tnnt3a | 0.472 | hmgb2b | -0.324 |
| casq2 | 0.470 | si:ch73-1a9.3 | -0.323 |
| cav3 | 0.469 | ran | -0.321 |
| srl | 0.450 | h2afvb | -0.317 |
| CABZ01072309.1 | 0.450 | khdrbs1a | -0.316 |
| ldb3b | 0.450 | fthl27 | -0.311 |
| pgam2 | 0.446 | cirbpb | -0.310 |
| ckmb | 0.436 | hnrnpaba | -0.307 |
| mustn1b | 0.436 | si:ch211-222l21.1 | -0.302 |
| atp2a1 | 0.431 | ptmab | -0.302 |
| trdn | 0.431 | ubc | -0.299 |
| desma | 0.430 | hnrnpabb | -0.296 |
| rtn2a | 0.430 | hmgb2a | -0.295 |
| ckma | 0.427 | cirbpa | -0.295 |
| calm3a | 0.426 | hmgn2 | -0.293 |
| ank1a | 0.422 | actb2 | -0.286 |
| pygma | 0.419 | si:ch73-281n10.2 | -0.285 |
| CABZ01078594.1 | 0.418 | cfl1 | -0.282 |
| tgm2a | 0.417 | hmga1a | -0.281 |
| myom1a | 0.417 | hmgn7 | -0.281 |
| tmod4 | 0.415 | syncrip | -0.281 |
| neb | 0.415 | hnrnpa0b | -0.278 |
| eno3 | 0.414 | setb | -0.278 |
| tmem38a | 0.412 | cbx3a | -0.277 |
| si:ch211-266g18.10 | 0.411 | tuba8l4 | -0.269 |
| klhl43 | 0.411 | sumo3a | -0.265 |
| gapdh | 0.410 | h2afva | -0.264 |
| aldoab | 0.410 | si:ch211-288g17.3 | -0.263 |
| phex | 0.407 | anp32b | -0.261 |
| sar1ab | 0.407 | anp32a | -0.260 |
| si:ch211-156j16.1 | 0.406 | snrpf | -0.259 |
| ak1 | 0.406 | snrpd1 | -0.255 |