"ligase III, DNA, ATP-dependent"
ZFIN 
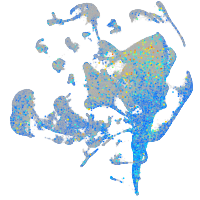
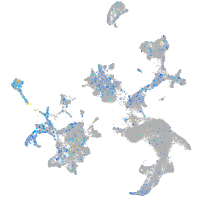
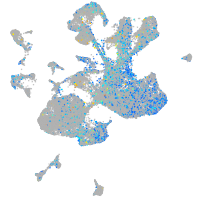
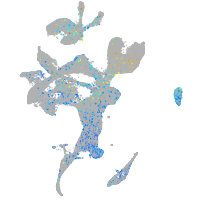
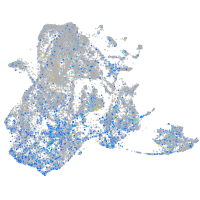
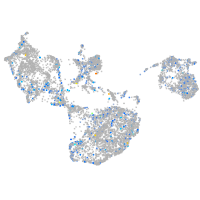
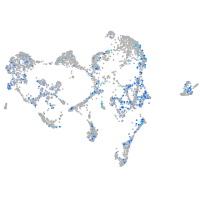
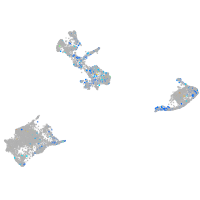
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmga1a | 0.157 | atp6v0cb | -0.108 |
| hspb1 | 0.153 | gapdhs | -0.107 |
| stm | 0.146 | sncb | -0.104 |
| si:ch211-152c2.3 | 0.145 | ptmaa | -0.102 |
| hnrnpa1b | 0.145 | ywhag2 | -0.100 |
| hnrnpub | 0.145 | cspg5a | -0.096 |
| cx43.4 | 0.141 | calm1b | -0.091 |
| rbm4.3 | 0.141 | gpm6ab | -0.091 |
| khdrbs1a | 0.139 | tpi1b | -0.091 |
| top1l | 0.139 | stmn2a | -0.090 |
| polr3gla | 0.138 | rnasekb | -0.090 |
| pou5f3 | 0.138 | atpv0e2 | -0.089 |
| seta | 0.136 | calm1a | -0.089 |
| hnrnpabb | 0.135 | sv2a | -0.088 |
| fbl | 0.133 | sypb | -0.088 |
| nop58 | 0.132 | snap25a | -0.088 |
| apoeb | 0.132 | zgc:65894 | -0.086 |
| npm1a | 0.131 | pvalb1 | -0.086 |
| ilf3b | 0.131 | rtn1b | -0.086 |
| ncl | 0.131 | vamp2 | -0.085 |
| srsf1a | 0.130 | stx1b | -0.084 |
| nop56 | 0.130 | pvalb2 | -0.084 |
| syncrip | 0.129 | gpm6aa | -0.083 |
| dkc1 | 0.129 | gng3 | -0.083 |
| snrpb | 0.128 | elavl4 | -0.082 |
| bms1 | 0.127 | atp6v1e1b | -0.082 |
| cirbpa | 0.127 | atp6ap2 | -0.082 |
| eif5a2 | 0.126 | sncgb | -0.082 |
| anp32e | 0.125 | map1aa | -0.082 |
| s100a1 | 0.124 | slc6a1a | -0.081 |
| si:dkey-66i24.9 | 0.123 | syt1a | -0.080 |
| NC-002333.4 | 0.122 | stxbp1a | -0.080 |
| hmgb2b | 0.122 | actc1b | -0.080 |
| smc1al | 0.122 | tmsb2 | -0.079 |
| crabp2b | 0.122 | eno2 | -0.079 |